Expression and visualization of Green Fluorescent Protein (GFP) in Neurospora crassa
We report the first successful imaging of GFP expression in Neurospora crassa. GFP was expressed under the control of the heterologous ToxA promoter from Pyrenophora tritici-repentis in transformants carrying multiple or single copies of the GFP construct. GFP was also detected in ascospores but not during earlier stages of the sexual cycle.
______________________________________________________________________________
Heterologous expression of Green Fluorescent Protein (GFP) from the jellyfish Aequorea victoria was first accomplished in Escherichia coli and Caenorhabditis elegans (M. Chalfie et al., 1994, Science 263:802). GFP has quickly become one of the favorite protein markers in many experimental systems (M. Chalfie and S. Kain [Eds.], 1998, "Green Fluorescent Protein: Properties, Applications and Protocols", Wiley-Liss, 385 pp) including many fungi (for review see J. M. Lorang et al., 2001, Appl. Env. Microbiol. 67:1987). Nevertheless, GFP constructs were not successfully expressed in N. crassa. Various forms of GFP under the control of constitutive or inducible fungal promoters or Neurospora-GFP fusion proteins have been tried but not found to be stably expressed at levels required for imaging.
In our most recent attempt to adapt GFP for N. crassa, we tested a transcriptional fusion of a strong fungal promoter with a GFP derivative of increased brightness ("S65T"; R. Heim et al., 1995, Nature 373:663) and with the C+G-rich codon bias of humans and N. crassa (J. Haas et al., 1996, Curr. Biol. 6:315). pCT74 contains the E. coli hph gene as selectable marker and a transcriptional fusion of the Pyrenophora tritici-repentis ToxA promoter (L. Ciuffetti et al., 1997, Plant Cell 9:135) with the altered GFP, sGFP. sGFP (W. Chiu et al., 1996, Curr. Biology 6:325) is identical to commercially available eGFP (Clontech). Transformation of other filamentous fungi with pCT74 resulted in GFP expression (J.M. Lorang et al., 2001, Appl. Env. Microbiol. 67:1987).
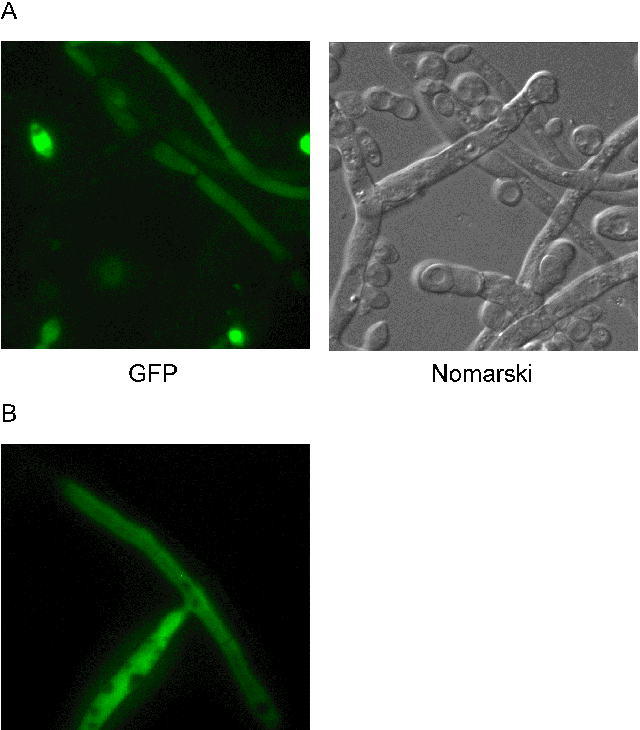
We transformed a N. crassa his-3 strain (FGSC# 6103) with pCT74 by electroporation (B.S. Margolin et al., 1997, FGN 44:34 and 2000, FGN 47:112). Transformants were selected on minimal medium supplemented with histidine and hygromycin B (Hyg; 200 ug/ml). Variable expression of GFP was detected by microscopy in macroconidia and hyphae of transformants carrying randomly integrated copies of ToxA-sGFP (Fig. 1A). Germinated conidia accumulated enough GFP in hyphae to be detected after overnight incubation at 32o C. Expression in young hyphae was strong and fairly uniform in most single copy transformants, whereas older hyphae showed lower fluorescence. GFP appeared to accumulate in macroconidia. As has been observed in plants (J. Haseloff and B. Amos, 1995, Trends Genetics 11:328), GFP appeared to accumulate in the nucleoplasm and to be excluded from vacuoles.
GFP expression levels and patterns differed from transformant to transformant, perhaps because of varied copy number, integration site and/or gene silencing. We therefore targeted single copies of sGFP to the his-3 locus (Fig. 1B). The resulting heterokaryotic transformants showed uniform and high-level expression of GFP.
To assess the expression of GFP in sexual tissues and the stability of GFP+ transformants during the sexual phase, we crossed single copy GFP+ strains to a wildtype strain (FGSC# 2490). GFP was expressed in protoperithicia and perithicia. Interestingly, GFP was not detected in dikaryotic and premeiotic tissue but was expressed in ascospores, regardless of whether GFP+ strains were used as females or males in heterozygous crosses (Fig. 2). GFP was detected even in fully melanized ascospores (Fig. 2C). Expression of GFP in hyphae was first seen at approximately 24 hours after ascospore germination. GFP was detected in all vegetative tissues and mature ascospores in GFP+ progeny from heterozygous crosses.
One explanation for past difficulties in creating strongly expressing Neurospora-GFP fusion proteins may be the relative strength of the promoters employed. The only published report of using a N. crassa promoter to drive GFP expression in fungi that we are aware of relates to experiments in Mycosphaerella graminicola, in which the acu-3 (isocitrate lyase) promoter region was used as a carbon source-repressed promoter (E.A. Rohel et al., 2001, Mol. Plant Microbe Interactions 14:156). In an attempt to find additional N. crassa promoters for GFP expression we performed a "promoter trap" screen. We co-transformed a N. crassa his-3 strain (FGSC# 6103) with a selectable marker (hph or his-3+) and various ToxA-sGFP fragments lacking a functional promoter. We screened approximately 10,000 transformants but were unable to find strains that stably expressed GFP without the full ToxA promoter. While our screen was not saturating, i.e. GFP could not have properly inserted in every promoter region of the N. crassa genome, it appears unlikely that this approach will result in the identification of strong promoters in N. crassa.
Previous failures to detect GFP expression may also be explained by gene silencing, for example by quelling and/or DNA methylation (E. Selker, 1997, Trends Genetics 13:296). We did not find any evidence for quelling of GFP in transformations with either pCT74 or ToxA-sGFP fragments. To explore the possibility that methylation was responsible for complete or partial silencing of GFP genes in N. crassa, we transformed N. crassa (FGSC# 6103) with his-3+ DNA from pNH60 (T.L. Legerton and C. Yanofsky, 1985, Gene 39:129) and DNA fragments with the promoterless sGFP (from pCT74) or jellyfish GFP (from pRSETB-GFP or pGFP10.1) gene and isolated twenty transformants carrying each construct. Surprisingly, we found that neither GFP gene alone was efficiently methylated in vegetative tissue of N. crassa (Fig. 3 and data not shown). Jellyfish GFP was predicted to be methylated because it has relatively high A+T content (62%) and a high TpA/ApT ratio (0.71; GenBank accession number M62653), features that mimic RIP-mutated N. crassa DNA which is usually methylated (V. Miao et al., 2000, J. Mol. Biol. 300:249). Modified sGFP and eGFP have low A+T content (39%) and a somewhat lower TpA/ApT ratio (0.67; accession number U43284) and thus were predicted to be unmethylated. In some transformants with ToxA-sGFP or promoterless sGFP some methylation was observed (e.g. TMF166-1, -5 and -8 in Fig. 3A) but presence of methylation did not always correlate with gene silencing. As expected, methylation was detected in sGFP genes that had undergone RIP (Fig. 3A, lanes 15 and 16) because sequences mutated by RIP can serve as efficient de novo methylation signals in vegetative tissues (M. Singer et al., 1995, Mol. Cell. Biol. 15:5586). We also found methylation in some multicopy pCT74 inserts that nevertheless expressed GFP at high levels (Fig. 3B). In these transformants, hph and plasmid fragments apparently promoted methylation, consistent with earlier results with chimeric plasmids (E. Selker et al., 1987, Science 238:48). These results encourage us to predict that transcriptional or translational fusions of fungal promoters or protein-coding regions to GFP will not be silenced by methylation in N. crassa.
Our results show that expression of GFP in N. crassa can be achieved by using transcriptional fusions of a strong fungal promoter and sGFP. Under our conditions, GFP had to accumulate for at least 16 hours in hyphae before visualization was possible. We created a set of GFP plasmids that allow sGFP targeting to the his-3 locus. It is unlikely that sGFP in future Neurospora-GFP fusion constructs will be silenced by methylation if single copies of genes are integrated into the genome by replacement strategies. Likewise, we have not found evidence for efficient quelling of GFP. GFP should immediately prove useful as an ascospore color marker, in genetic screens and for gene expression studies. Neurospora-GFP fusion proteins under construction in our lab will allow studies on the subcellular localization of organelles and proteins and will facilitate cell biology research with N. crassa.
Acknowledgements:
We would like to thank Chris Doe and Bruce Bowerman, both at the University of Oregon (UO), for the use of their laboratories' microscopes. M.F. would like to thank Aaron Severson (UO) and Sal Fuerstenberg (UO) for an introduction to imaging of GFP by microscopy. Rebekka Wachter (UO) provided pRSETB-GFP and Tim Bestor (Columbia Univ.) provided pGFP10.1, both plasmids containing jellyfish GFP cDNA. This study was supported by grants from the National Institutes of Health to M.F. (CA73123) and E.U.S. (GM35690).
Figure 1 (Following page): Expression of GFP in N. crassa. (A) GFP in 3-day old hyphae and macroconidia of a heterokaryotic primary transformant (TMF153-9) grown on minimal medium supplemented with Hyg. (B) Expression of sGFP was uniform in GFP+ His+ transformants in which ToxA-sGFP had been targeted to the his-3 locus (N2002). pMF245 (ToxA-sGFP without terminator) and pMF246 (ToxA-sGFP with the nos terminator), respectively, were created by inserting ApaI - NotI or ApaI - EcoRI fragments of pCT74 into the same sites of pBM61 (B.S. Margolin et al., 1997, FGN 44:34). As expected, we found that the nopaline synthase (nos) terminator did not affect expression of GFP in N. crassa (data not shown). To detect GFP microscopically, we used a Zeiss Axioscope with a 100X oil immersion Plan-Neofluar objective (N.A. 1.3 mm), a Zeiss filter set (green excitation filter 450-490 HB) and a Zeiss AttoArc2 HB-100W mercury lamp. Images were captured with a Micromax EBF512 camera (Princeton Instruments) and processed with the MetaMorph Imaging System (Universal Imaging Corp.) and Adobe Photoshop software. Typical exposure times were 5 - 15 ms for both GFP and Nomarski images. Color reproductions of figures shown will be made available with the on-line version of this article on the FGSC website (http://www.fgsc.net/).

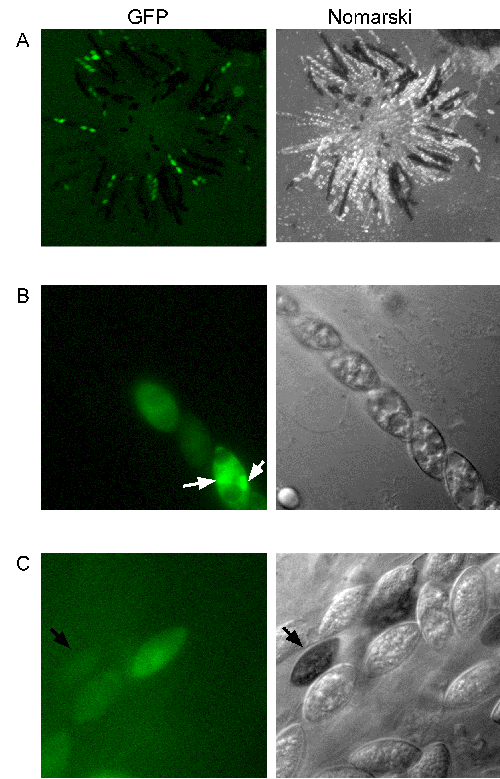 Figure 2: Expression of GFP in sexual tissues. (A) Perithicia from a heterozygous cross of the heterokaryotic GFP+ strain TMF153-9 (male) to FGSC #2490 (female) were squashed at approximately 10 - 14 days post fertilization to extrude developing asci and examined for GFP expression. Tissue in early stages of post fertilization development apparently did not express GFP. GFP+ / GFP- segregation patterns are evident. Images were captured as described in Fig.1, except that a 5X Plan-Neofluar objective (N.A. 0.15 mm) was used. Exposure times had to be increased to 100 ms; exposure times for the Nomarski images were approximately 10 ms. (B) Segregation of GFP- / GFP+ ascospores in one ascus. GFP appears to accumulate in nuclei (white arrows). (C) GFP can be detected even in melanized ascospores (black arrow). Images in (B) and (C) were captured as described in Fig. 1.
Figure 2: Expression of GFP in sexual tissues. (A) Perithicia from a heterozygous cross of the heterokaryotic GFP+ strain TMF153-9 (male) to FGSC #2490 (female) were squashed at approximately 10 - 14 days post fertilization to extrude developing asci and examined for GFP expression. Tissue in early stages of post fertilization development apparently did not express GFP. GFP+ / GFP- segregation patterns are evident. Images were captured as described in Fig.1, except that a 5X Plan-Neofluar objective (N.A. 0.15 mm) was used. Exposure times had to be increased to 100 ms; exposure times for the Nomarski images were approximately 10 ms. (B) Segregation of GFP- / GFP+ ascospores in one ascus. GFP appears to accumulate in nuclei (white arrows). (C) GFP can be detected even in melanized ascospores (black arrow). Images in (B) and (C) were captured as described in Fig. 1.
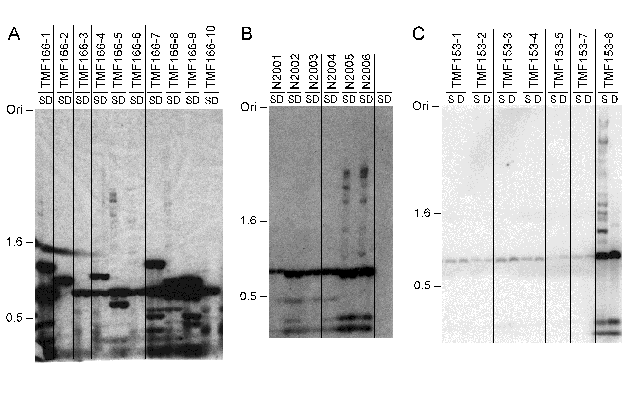
Figure 3: DNA methylation analysis of selected sGFP+ transformants. (A) Southern analysis of genomic DNA isolated from transformants with randomly inserted copies of sGFP showed that sGFP remained mostly unmethylated (TMF166-1 to -10). Genomic DNA of sGFP+ transformants was digested with methylation-sensitive (Sau3AI; S) or methylation-insensitive (DpnII; D) isoschizomers, fractionated in agarose gels, blotted to nylon membranes and probed with a radioactively labeled sGFP fragment. (B) Single copies of sGFP integrated at his-3 were always unmethylated (N2001 to N2004), whereas RIP-mutated alleles of sGFP were methylated (N2005 and N2006); lane H shows the non-transformed host strain (FGSC# 6103). (C) SGFP was unmethylated in transformants (TMF153-1 to -7) with single copies of pCT74, but methylated in a transformant (TMF153-8) with multiple copies of the plasmid. GFP was expressed at high levels in all seven strains. The position of size markers (kb) and the gel origin (Ori) are indicated.