The Neurospora crassa Mitochondrial Genome*
*Based on Mitochondrial genetics of Neurospora , a review by A J. F. Griffiths, R. A. Collins, and F. E. Nargang ,
in "The Mycota II, Genetics and Biotechnology" (U. Kück, Ed.), pp. 93-105. Springer-Verlag. Berlin, Heidelberg, 1995.
See also Hudspeth, M. E. S., 1992 The fungal mitochondrial genome—a broader perspective, in "Handbook of Applied
Mycology. Volume 4: Fungal Biotechnology" ( D. K Arora, R. P. Elander, and K. G. Mukerji, eds.), pp. 213-241.
Marcel Dekker, New York.
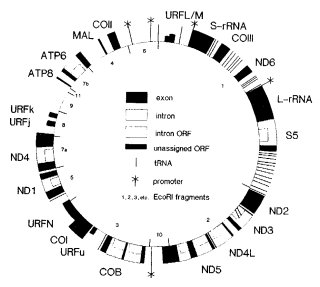
Appendix 4 Figure 1 Physical map of the mitochondrial DNA of wild type strain 74-OR23-1A. The corresponding genes are
identified in Appendix 6 Table 1. For a map of restriction endonuclease cleavage sites of the same strain, see Taylor and
Smolich (Curr. Genet. 9:597-603 1995). From Griffiths, Collins and Nargang (1995), with permission from Springer-Verlag..
APPENDIX 4: TABLE 1
Mitochondrial Genes of Neurospora crassa
___________________________________________________________________________________________________
Genea Equivalentb Encoded Product
___________________________________________________________________________________________________
Respiratory chain
cox1 coI Cytochrome c oxidase subunit 1
cox2 coII Cytochrome c oxidase subunit 2
cox3 coIII Cytochrome c oxidase subunit-3
cob cob Apocytochrome b
ndh1 ND1 NADH dehydrogenase subunit 1
ndh2 ND2 NADH dehydrogenase subunit 2
ndh3 ND3 NADH dehydrogenase subunit 3
ndh4 ND4 NADH dehydrogenase subunit 4
ndh4l ND4L NADH dehydrogenase subunit 4L
ndh5 ND5 NADH dehydrogenase subunit 5
ndh6 ND6 NADH dehydrogenase subunit 6
ATP synthesis
atp6 ATP6 ATP synthase subunit 6
atp8 ATP8 ATP synthase subunit 8
atp93 MAL (ATP synthase subunit 9)3
Translational apparatus
rnl L-rRNA Large ribosomal subunit RNA
rns S-rRNA Small ribosomal subunit RNA
S-5 S5 Small ribosomal subunit protein
tsld tRNA tRNA
Unidentified reading frame
URF URF j
URF k
URF L/M
URF N
URF u
Open reading frame
ORFe ORF
___________________________________________________________________________________________________
a Symbols used for fungal mitochondrial genes by Hudspeth 1992 and recommended for Neurospora.
b Symbols used for Neurospora mitochondrial genes in Figure 1 and in Griffiths et al. 1995.
c The Neurospora atp9 gene is translationally inactive. The translationally active gene for ATP synthase subunit 9 is the nuclear gene
oli, which is closer in sequence to the translationally active mitochondrial gene ATP9 of Saccharomyces.
d "tRNA synthesis locus" (Hudspeth 1992).
e Several intron-encoded ORFs have been identified.
APPENDIX 4: TABLE 2
Mutant Mitochondrian Genomes of Neurospora crassaa
| Group |
Mutant |
Growth |
Cytochromes |
Suppression by
su([mi-1])-f su([mi-1])-1 |
|
| I |
[poky](=[mi-1]) |
Lag |
Low aa3, and b, high c |
+ - |
| |
[SG-1,-3] |
Lag |
Low aa3, and b, high c |
+ - |
| |
[stp-B1] |
Lag |
Low aa3, and b, high c |
+ - |
| |
[exn-1,2,3,4] |
Lag |
Low aa3, and b, high c |
+ - |
| |
[C93] |
Temperature Sensitive |
Low aa3, and b, high c |
? ? |
| II |
[mi-3] |
Less Lag |
Low aa3, high b, normal c |
- + |
| |
[exn-5] |
Loss Lag |
Low aa3, high b, normal c |
- + |
| III |
[stp] |
Stop-start |
Low aa3, and b, high c |
- - |
| |
[stp-A, -A18,-B2, -C] |
Stop-start |
Low aa3, and b, high c |
- - |
| |
[abn-1, -2] |
Stop-start |
Low aa3, and b, high c |
- - |
| |
[E35] |
Stop-start |
? |
? ? |
| |
[ER-3] |
Stop-start |
? |
? ? |
| |
[stp-107] |
Stop-start |
Low b and c, no aa3 |
? ? |
aThe mutations include nucleotide-pair substitutions, deletions, duplications,
insertions, and complex rearrangements. The strains with mutant mitochondrial
are cyanide-insensitive and salicyl hydroxamide-sensitive. They must thus rely
on an alternate oxidase system. Except for Group II, they are all defective in
mitochondrial protein synthesis. Inclusion of [C93] in Group I is tentative.
Mutant names: SG, slow growth; stp, stopper: exn, extranuclear; mi, maternally
inherited; abn, abnormal growth. Adapted from a table of Griffiths et al. 1995
and based on references given in that review.
Return to the 2000 Neurospora compendium main page
Return to the FGSC home page
