These one-letter symbols are used in most contexts as abbreviations for mat A and mat a, designating idiomorphs at the mating-type locus in linkage group I.
aab-1 : am α-binding protein-1
IIIR. Linked to trp-1 and con-7 by RFLP (370).
Cloned and sequenced: EMBL/GenBank AF026550.
Specifies subunit of a nuclear heteromeric complex that binds to a CCAAT box in the upstream activating sequence of am. AAB-1 is similar to Saccharomyces cerevisiae HAP5. Disruption of aab-1 by RIP not only cuts in half the level of glutamate dehydrogenase but also affects growth and development adversely, resulting in short aerial hyphae and reduced conidiation. This indicates that other genes in addition to am are subject to regulation by aab-1 (370).
aac : ADP/ATP carrier protein
Unmapped.
Cloned and sequenced: Swissprot ADT_NEUCR, EMBL/GenBank X00363, PIR A03182, XWNC, GenBank NCADPATP.
Encodes ADP/ATP carrier protein (translocase), adenine nucleotide translocator, exchanging ADP and ATP across the mitochondrial inner membrane (50). Called acp.
aaf : acetylaminofluorine requirement
Unmapped. The data are said to be consistent with one gene.
Complex phenotype. The requirement is satisfied by 2-acetylaminofluorine, certain azo dyes, or certain single amino acids. Cold sensitive. Found among progeny of a rib-1 strain that had become tolerant to 2-acetylaminofluorine (2080).
aag-1 : accelerated acetate growth
II. Linked to arg-5 (33%), acu-5 (26%) (367).
Growth is faster than wild-type on acetate as the sole carbon source and slower on sucrose as the sole carbon source. Derepressed, with significant constitutive levels of acetyl-CoA synthetase and glyoxylate cycle enzymes on sucrose; semidominant (367).
aap-2 : amino acid permease-2
IR. Linked to al-2 (0T/18 asci) (1447).
Cloned and sequenced: EMBL/GenBank AF053231.
Encodes a polypeptide predicted to have 12 membrane-spanning regions. The predicted AAP-2 polypeptide has significant sequence similarity to GABA transport proteins in other organisms; its function in Neurospora crassa is unknown (1270).
ace : acetate
Acetate mutants ace-1 through ace-7 are auxotrophs that grow on 0.3% sodium acetate, as do suc mutants (which often grow better on acetate than on succinate). A carbon source is also needed. Most ace mutants grow better when the carbon source is maltose rather than sucrose (1122). Acetate mutants, except ace-1 and ace-5, can grow on various Tweens as the sole carbon source (232). The mutants differ in their ability to grow on complex media. Unlinked genes ace-2, -3, and -4 are involved with the pyruvate dehydrogenase complex (1496), a complex enzyme that in yeast is formed from polypeptides encoded by three different genes, with the activities pyruvate dehydrogenase (E1) (EC 1.2.4.1), dihydrolipoamide acetyltransferase (E2) (EC 2.3.1.12) and lipoamide dehydrogenase (E3) (EC 1.8.1.4). A separate set of acetate mutations called ac-1, -2, -3, -4, and -5 (2011) was lost before being mapped or tested for allelism with the mutants now called ace-1 through ace-7.
ace-1 : acetate-1
IIR. Between un-20 (15%) and eas (1%), fl (11%) (103, 1546).
Requires acetate. Poor growth on complex complete medium. Grows well on acetate (0.1%) aided by ethanol (0.5%) [E. L. Tatum and L. Garnjobst, cited in refs. (103) and (1582). Ascospore maturation and germination are slow. Germination is best on sucrose minimal medium with yeast extract and ethanol (714). Not the same as ac-1 of ref. (2011), which was lost. Called ac.
ace-2 : acetate-2
IIIR. Between pro-1 (1%; 9%; 36 kb) and com (5%), ad-4 (4%; 7%), met-8 (448, 1122, 1546).
Cloned and sequenced: Swissprot ODP2_NEUCR, EMBL / GenBank J04432, EMBL NCPNUC, GenBank NEURPNUC; pSV50 clones 1:6G, 11:1B, 18:12G; pCRD129, pCRD130 (448).
Requires acetate. Will not use succinate or ethanol (556). Encodes the dihydrolipoamide acetyltransferase (EC 2.3.1.12) component (E2) of pyruvate dehydrogenase complex EC 1.2.4.1 (1496). Good growth on complex complete medium (1122). Not the same as ac-2 of ref.erence (2011).
ace-3 : acetate-3
IR. Between lys-3 (£1%; 1%), In(OY323)R and nic-1 (<1%). Included in duplications from In(OY323) ´ In(NM176) (9, 115, 1122, 1971).
Cloned / partially sequenced candidates: Either GenBank AI399057 (EST W17H2) or AI397651 (EST SC5E2), which show homology to the a- and b-subunits of the pyruvate dehydrogenase E1 component, respectively (1448).
Requires acetate. Lacks pyruvate dehydrogenase complex activity (EC 1.2.4.1) (1496). Grows poorly on complex complete medium (1122). Conidiation is best at 25°C, not 34°C (1546). Not the same as ac-3 of ref.. (2011).
ace-4 : acetate-4
IVL. Between cys-10 (19%; 27%) and fi (10%) (1122).
Cloned / partially sequenced candidates: Either GenBank AI399057 (EST W17H2) or AI397651 (EST SC5E2), which show homology to the a- and b-subunits of the pyruvate dehydrogenase E1 component, respectively (1448).
Requires acetate. Grows on complex complete medium (1122). Lacks pyruvate dehydrogenase complex activity, (EC 1.2.4.1) (1496). Lipoate acetyltransferase fails to aggregate to form the core of the pyruvate dehydrogenase complex. As a result, there is high activity of the free components pyruvate dehydrogenase and lipoamide reductase (1496). Not the same as ac-4 of ref. (2011).
ace-5 : acetate-5
VR. Between gul-1 (<1%) and ure-1 (<1%) (1121, 1122).
Requires acetate. Grows poorly on complex complete medium (1122). Not the same as ac-5 of ref. (2011).
ace-6 : acetate-6
Name not used because suc has priority. See suc.
ace-7 : acetate-7
IR. Between nic-2 (4%; 7%) and cr-1 (1%; 3%) (1122).
Requires acetate. Structural gene for glucose-6-phosphate dehydrogenase (G6PDH) (EC 1.1.1.49); the enzyme in two revertants is qualitatively different from wild-type G6PDH. Pyruvate dehydrogenase and pyruvate carboxylase activities are normal (1122). Grows well on complex complete medium. Unable to use xylose as a carbon source, resembling suc mutants and differing from the wild type and all other ace mutants in this respect.
ace-8 : acetate-8
VIIL. Between thi-3 (1%), T(T54M50) and qa-3 (2% or 3%) (1120, 1578).
Structural gene for pyruvate kinase (EC 2.7.1.40) (1123). Growth and conidiation are best on acetate plus ethanol or ethanol plus L-alanine.
ace-9 : acetate-9
IIR. Between nuc-2 (2%) and arg-12 (3%), aro-1 (10%) (1788, 1789).
Requires acetate. Very weak activity of the pyruvate dehydrogenase complex. Grows well on complex medium (1788, 1789).
acon-1 : aconidiate-1
Allelic with fl.
acon-2 : aconidiate-2
IIIR. Linked to vel (6%), tyr-1 (9%; 14%), probably between them (1293, 1582)
Macroconidiation is defective. Allele RS91 is heat sensitive, with macroconidiation blocked early in the developmental pathway at 34ºC. No minor constriction budding. Scanning EM photograph (1958). Some conidia are formed at 25ºC, but growth is subnormal (1293). Homozygous fertile (1582).
acon-3 : aconidiate-3
IVL. Between cys-10 (1%; 6%) and cut (33%) (1582). A report of linkage in VIL was not confirmed.
Macroconidiation is blocked (1293). No major constriction budding. Scanning EM photograph (1958). Some conidia have been observed low in slants at 25ºC. Female sterile (1582).
acp-1 : acyl carrier protein
Unmapped.
Cloned and sequenced: Swissprot ACPM_NEUCR, EMBL/GenBank X83578, NCMTACP, PIR S00491, S17647 (1825).
Encodes acyl carrier protein precursor (ACP), NADH-oxidoreductase 9.6-kDa subunit (EC 1.6.5.3, EC 1.6.99.3). Mitochondrial acyl carrier protein (240) is part of complex I (1774, 1825), where it has a role in both lipid metabolism and complex formation [ 1824 , 1825, 2149; see nuo in ref. (2299)]. Called nuo9.6 (1843). The symbol acp was used temporarily for genes specifying ADP / ATP carrier protein (aac) and inducible acetate permease (acu-16).
acpi : inducible acetate permease
Changed to acu-16.
acr-1 : acriflavine resistant-1
IL. Linked to mat (8%; 12%) (931).
Shows low-level resistance to acriflavine (2 ug/ml on agar medium) (931). Scoring may be difficult with some tester strains because differences in sensitivity probably exist between different laboratory strains, with wild-type STA4 being less sensitive to acriflavine than is wild-type Pa, which was used in the original study (1546).
acr-2 : acriflavine resistant-2
III. Between acr-7 and T(T54M140b), thi-4 (0/286), sc (3%; 6%), spg (1%; 11%) (931, 1546, 1578, 1592). acr-2 and trp-1 (IIIR) cosegregated at the second division in 1 of 13 asci (914), which would favor a right-arm location for acr-2. However, acr-2 is in IIIL if the T(OY339) breakpoint is left of Cen-III (1546).
Cloned and sequenced: EMBL/GenBank D45893, PIR S72537, S78458 for the main open reading frame, plus the two short upstream open reading frames PIR S72535 and S72536; cosmid pDC107 (27).
Mutant alleles obtained by selecting for resistance to acriflavine (927, 928) are resistant to 3-AT (927) but not to acridine orange, acridine yellow, or malachite green (27). They are also not cross-resistant to cycloheximide and oligomycin, unlike pleiotropic drug-resistant (PDR) mutants of Saccharomyces. An excellent stable marker, fully fertile. Small inocula should be used to avoid false-positive tests (280). Acriflavine is used at 50 ug/ml in minimal agar medium (1592) (higher concentrations may be used), or 3-AT at 0.5 mg/ml, both added before autoclaving. Resistance is dominant (27, 931). For cloning, a library was therefore constructed using resistant mutant KH2, in which an asparagine residue is changed to lysine. Disruption of acr-2+ by RIP results in greater sensitivity than that of wild type. Resistance is thus due to functional change in the ACR-2 protein rather than to loss of function of the protein (27). The gene includes two small upstream open reading frames, which are probably regulatory (27).
acr-3 : acriflavine resistant-3
IL. Between un-16 (1%; 5%) and suc (1%; 5%), In(H4250)L. Probably right of ta (1582, 1592).
Resistant to acriflavine and to malachite green, but not to 3-AT, on agar medium (931). Reported not resistant to malachite green in liquid medium, or to acridine orange, acridine yellow, or 3-AT (27). Resistance is dominant in duplications from In(H4250) and T(39311) (1546) and in heterokaryon tests (931). Scoring on agar is clear when uniform inocula of appropriate size are used, but false-negative or false-positive scoring may result if test inocula are too small or too large. Tests should be read at 2 and 4 days (34°C) to detect possible delayed expression of resistance. On minimal agar medium at 34°C, acriflavine is used at 10 ug/ml (1592) and malachite green at 2 ug/ml (931).
acr-4 : acriflavine resistant-4
IL. Linked to mat, acr-3 (5%) (932).
Resistant to acriflavine (50 ug/ml) when acr-4 is combined with the morphological mutation shg (932). Also resistant to acridine orange and acridine yellow, but not to 3-AT or malachite green (27).
acr-5 : acriflavine resistant-5
IIR. Between arg-5 (6%) and pe (9%) (1593).
Resistant to acriflavine (50 ug/ml) when acr-5 allele KH16 is combined with morphological mutation mo(KH161) (932). Allele JLC74, however, is readily scorable in strains of wild type morphology (1546). Not resistant to 3-AT, acridine orange, acridine yellow, or malachite green (27).
acr-6 : acriflavine resistant-6
IIIR. Linked to shg (0/368) (932).
Resistant to acriflavine (50 ug/ml). The shg strain in which acr-6 originated is acriflavine-sensitive (932). Also resistant to acridine orange and acridine yellow, but not to 3-AT or malachite green (27).
acr-7 : acriflavine resistant-7
IIIL. Between r(Sk-2)-I (7%), T(NM183) (5%) and acr-2 (12%), thi-4 (7%), sc (12%; 14%) (280, 1582, 2121). A report of VI linkage in ref. (1603) is incorrect.
Resistant to acriflavine (50 ug/ml). Not resistant to 3-AT, malachite green, acridine orange, or acridine yellow (27, 1582). Several acr-7 strains have become female-infertile after vegetative transfer (1582). In the presence of cum, scoring of acr-7 requires very small inocula that are best obtained by floating conidia down onto agar medium without touching the surface (2121).
act : actidione resistant
Changed to cyh.
act : actin
VR. Linked near inl (2092).
Cloned and sequenced: Swissprot ACT_NEUCR, EMBL/GenBank U78026.
Structural gene for actin. Filamentous actin is localized primarily to hyphal tips. Transcript levels decrease on induction of conidiation (2092). The symbol act was once used for mutants resistant to cycloheximide (then called actidione). These were then renamed cyh: cycloheximide-resistant. The symbol act is retained here for the actin gene. Doing so violates priority, but is less likely to confuse than would be the creation of a new symbol for actin. For an actin-related gene, see arp3.
acu: acetate utilization
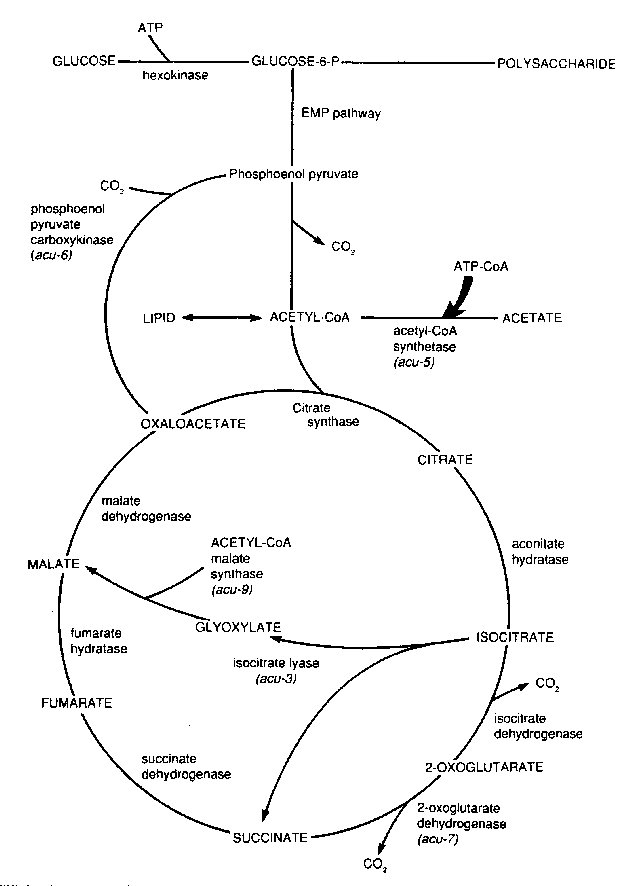
Unable to use acetate as a carbon source. When ammonium acetate (3 mg/ml) is the sole carbon source, the wild type shows sparse but positive growth, in contrast to clear blanks for acu mutants. Mutants are selectable by inositol-less death on acetate medium. acu-1, acu-5, acu-6, and acu-7 do not behave as respiratory mutants in tetrazolium-overlay tests on acetate medium (593). Positions of mutants in the metabolic pathway are shown in Fig. 4.
acu-1 : acetate utilization-1
VR. Right of asn (21%) (651).
Unable to use acetate as a carbon source (651, 652). Selected by inositol-less death on acetate medium.
acu-2 : acetate utilization-2
IVR. Between leu-2 (11%) and pan-1 (6%) (651).
Unable to use acetate as a carbon source. Reduced level of oxoglutarate dehydrogenase (EC 2.3.1.61). Poor recovery from ascospores (651, 652).
acu-3 : acetate utilization-3
VR. Between inl (7%) and asn (20%) (1149).
Cloned and sequenced: Swissprot ACEA_NEUCR, PIR S26858, S22057, EMBL/GenBank X62697, GenBank NCACU3; EST NM3F4.
Structural gene for isocitrate lyase (EC 4.1.3.1). Unable to use acetate as a carbon source. Some revertants produce a temperature-sensitive enzyme (1149) (Fig. 4).
acu-4 : acetate utilization-4
IR. Right of arg-1 (5/29) (651).
Unable to use acetate as a carbon source (651, 652).
acu-5 : acetate utilization-5
IIR. Right of arg-5 (6%), aro-3 (7%) (651).
Cloned and sequenced: Swissprot ACSA_NEUCR, PIR S09244, SYNCAA, EMBL/GenBank X16989, Z47725, GenBank NCACU5, NCACCOASY.
Unable to use acetate as a carbon source. Structural gene for acetyl coenzyme A synthetase (EC 6.2.1.1) (acetyl-CoA ligase, acyl-activating enzyme) (652). See Figs. 4 and 17.
acu-6 : acetate utilization-6
VIL. Left of cys-1 (3%) (143, 651).
Unable to use acetate as a carbon source (651). Structural gene for phosphoenolpyruvate carboxykinase (EC 4.1.1.31) (143, 652) (Fig. 4). Strains with some complementing alleles possess protein that is electrophoretically similar to the enzyme. A temperature-sensitive partial revertant enzyme maps at the original locus (143). Interallelic complementation (651).
acu-7 : acetate utilization-7
IIIR. Linked to dow (0/72) (1582).
Unable to use acetate as a carbon source. Recovery from ascospores is poor (~25%) (651). Reduced level of oxoglutarate dehydrogenase (EC 2.3.1.61) (652) (Fig. 4).
acu-8 : acetate utilization-8
IIR. Between trp-3 (8%) and un-15 (3%) (1263).
Cloned and sequenced: Swissprot ACU8_NEUCR, PIR A36316, EMBL/GenBank M31521, GenBank NEUACU8, EST NC4G6.
Structural gene for acetate permease (1263)

FIGURE 4 The tricarboxylic acid cycle and the anaplerotic glyoxylate cycles showing acu mutants affected in acetate utilization. Not shown is acu-8, specifying acetate permease. From ref. (406), with permission from Wiley-VCH Verlag.
acu-9 : acetate utilization-9
VII. Linked to nic-3 (29%) (405).
Cloned and sequenced: Swissprot MASY_NEUCR, PIR S17774, EMBL/GenBank X56627, GenBank NCACU9.
Structural gene for malate synthase (glyoxysomal) (EC 4.1.3.2) (1784) (Fig. 4). Mutant obtained by RIP (405).
acu-10 : acetate utilization-10
Unmapped.
Resistant to fluoroacetate (1512).
acu-11 : acetate utilization-11
VII. Linked to arg-10 (21%) (1512).
Resistant to fluoroacetate (1512).
acu-12 : acetate utilization-12
IIR. Linked to trp-3 (24%) (1512).
Resistant to fluoroacetate (1512).
acu-13 : acetate utilization-13
IIR. Linked to trp-3 (23%) (1512).
Resistant to fluoroacetate. No growth on ethanol (1512).
acu-14 : acetate utilization-14
VI? Perhaps loosely linked to ylo-1 (404).
Does not complement any other acu mutant. There is no clear enzyme defect, but the levels of acetyl-CoA synthase and isocitrate lyase are affected (367).
acu-15 : acetate utilization-15
Unmapped.
Cloned and sequenced: Swissprot AC15_NEUCR, EMBL/GenBank Y11565, GenBank NC11565.
Encodes acetate metabolism transcriptional activator; a zinc-finger protein that binds upstream of acu genes (178). Mutants isolated through RIP show a reduction in growth on acetate. Unlike the facB mutants of Aspergillus, they are not appreciably more resistant to fluoroacetate than is wild type. When transferred to acetate growth medium, they do, however, show weak induction of acetyl-CoA synthetase and poor induction of the glyoxylate enzymes (404).
acu-16 : acetate utilization-16
Unmapped.
Not allelic with other acu mutations. Described as lacking an inducible acetate transport system (1696). More recently found to take up acetate into the mycelium, but with no apparent glyoxalate cycle flux (406, 2082). Originally called acpi : inducible acetate permease, but renamed because characterization of its function has been revised and because the symbol acp is used for a gene specifying acyl carrier protein (240).
ad: adenine
For the purine biosynthetic pathway, see Fig. 5. ad-5 catalyzes two sequential steps in the pathway, and, by analogy with yeast, the gene specifies a single polypeptide with two enzymatic activities. ad-1 appears to be an isogene. A similar situation pertains in yeast, adea16 and ade17 being the isogenes for these two steps. ad-4 carries out two lyase reactions on different but structurally related substrates at different steps in the pathway. ad-2 has until now been regarded as specifying the fifth step in the adenine pathway, but its homolog in yeast also specifies the second step. Growth of mutants in terminal (post-AICAR) steps of the adenine biosynthetic pathway is aided by histidine, which has a sparing effect on ad-l, ad-4 and ad-8 (295, 1314). Some ad-5 alleles are aided by histidine, others inhibited (295). Mutants of ad-3B, ad-4, ad-8, and ad-9 have been used to study the effect of histidine on purine pool utilization (1543). Mutants at the ad-3A and ad-3B loci accumulate purple pigment when adenine is limiting. Smaller amounts of the pigment may be seen in other post-AIR genes such as ad-4 and ad-5 (997, 1360). The pigment may consist of polymerized AIR (1543). Mutants affecting earlier biosynthetic steps are epistatic to ad-3 and later mutants with respect to purple pigment production (997). ad-3B (and presumably also ad-3A) cultures accumulate spontaneous mutants at other ad loci; these prevent pigment production and improve the growth rate of the double mutant (1368). Mutants of ad-3B, ad-4, ad-8, and ad-9 have been used to study the effect of histidine on purine pool utilization (1543). For regulation of purine catabolism, see the review in ref. (1281). For regulation of purine biosynthesis, see refs. (767) and (1545). For interrelation of purine, histidine, and tryptophan pathways, see ref. (1543). Indole may strongly inhibit adenine mutants (1159). Adenine mutants at the various loci were originally assigned to complementation groups designated by capital letters (997). Relation of most of these groups to steps of the biosynthetic pathway are given in Fig. 10 of ref. (249).
ad-1 : adenine-l
VIL. Between ylo-1 (6%) and Cen-VI (l% or 2%), T(AR209), glp-4 (0; 2%), rib-1 (3%; 5%) (1986, 2160).
Uses adenine or hypoxanthine (1360, 1620). Accumulates AICAR (160, 1764) and SAICAR(160). By analogy with yeast, a putative isogene which, with ad-5, specifies AICAR transformylase (EC 2.1.2.3) and IMP cyclohydrolase (EC 3.5.4.10) (Fig. 5). Used to study purine transport (1765). Ascospores are white and inviable in homozygous ad-1 x ad-1 crosses, and the ad-1 ascospores may be white in heterozygous crosses (1546). Called complementation group M.
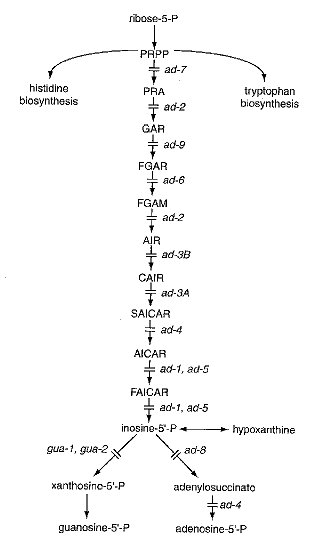
FIGURE 5 The purine biosynthetic pathway and sites of action of ad and gua genes (160, 249, 650, 736, 767, 964, 996). Abbreviations: PRPP, 5-phosphoribosyl pyrophosphate; PRA, 5-phosphoribosylamine; GAR, 5 -phosphoribosylglycineamide; FGAR, N-5 -phosphoribosylformylglycineamide; FGAM, 5 -phosphoribosylformylglycineamidine; AIR, 5 -phosphoribosyl-5-aminoimidazole; CAIR, 5 -phosphoribosyl-5-aminoimidazole-4-carboxylate; SAICAR, 5 -phosphoribosyl-5-aminoimidazole-4-N-succinocarboxamide; AICAR, 5 -phosphoribosyl-5-aminoimidazole-4-carboxamide; FAICAR, 5 -phosphoribosyl-5-formamidoimidazole-4-carboxamide. From ref. (1596), with permission from the American Society for Microbiology.
ad-2 : adenine-2
IIIR. Between thi-2 (1%) and trp-1 (1%; 7%) (18, 426).
Requires adenine or hypoxanthine (1360). The mutant is defective in phosphoribosylglycinamide cycloligase (AIR synthetase) (EC 6.3.3.1) (249), which catalyzes the fifth step in purine biosynthesis (Fig. 5). The yeast homolog is bifunctional, also specifying the phosphoribosylamine-glycine ligase (EC 6.3.4.13), the second step in the pathway, for which no gene has been identified in Neurospora. Allele 70004(t) is heat-sensitive (34 vs 25ºC) (1360) and osmotic-remediable (1273). Called complementation group H.
ad-3A : adenine-3A
IR. Between ure-4, his-3 (1% or 2%) and ad-3B (<1%) (156, 492).
Cloned and partially sequenced: EMBL/Genbank AI416404; EST NC2G7.
Requires adenine or hypoxanthine (1360). Blocked in the interconversion of CAIR + aspartate to SAICAR (SAICAR synthase, EC 6.3.2.6) (650) (Fig. 5). Produces purple pigment, permitting direct visual selection. Pigment is secreted with low adenine supplement (e.g., 0.l mM), not with high (2 mM) (503, 1360, 1542). Fertility is reduced in crosses between alleles (771). There is no interallelic complementation (486, 1896). Either forward mutation (504) or reverse mutation (1500) can be measured precisely. Production of purple pigment permits visual detection of mutants (650). ad-3A mutants N23 and N24 have been used extensively as mutagen testers: N23 reverts with agents that cause base-pair substitutions, and N24 reverts with agents that cause frame shifts (1500). Pigment production has been used to assess the effect of histidine and tryptophan on purine nucleotide synthesis (1543). SK(ad-3A) is a mutation at or near ad-3A and may be a cryptic ad-3A allele. It does not require adenine for growth. In crosses of SK(ad-3A) ´ ad-3A, the ad-3A progeny die. Possibly SK(ad-3A) fails to make enough adenine to support their growth (516). Translocations T(Y155M64) ad-3A (493, 1582) and T(Y112M15) ad-3A (774) each have one breakpoint inseparable from ad-3A. The letter A in the locus symbol does not refer to mating type; it was appended because ad-3A mutants were originally called complementation group A (487).
ad-3A and ad-3B are two genetically and functionally distinct loci separated by a short but functionally complex region of unknown but essential function (492, 771). They have been used to develop an ad-3A and ad-3B specific-locus assay system for quantitative genetic and molecular studies of mutation [see introduction to ref. (497)]. The marked ad-3A + ad-3B heterokaryons used in the assay system shelter not only mutations in single genes but also recessive lethal multilocus deletions, enabling these to be characterized. The system has been used in massive studies with various physical and chemical agents (495-497, 499, 500, 505, 506, 827). Characterized mutations that were induced by various agents are available from FGSC.
ad-3B : adenine 3B
IR. Between ad-3A (<1%) and T(P7442)L, nic-2 (3%) (492).
Uses adenine or hypoxanthine (1360). Blocked in the interconversion of AIR to CAIR (AIR carboxylase, EC 4.1.1.21) (650) (Fig. 5). Produces purple pigment, permitting direct visual selection. Pigment is secreted with low adenine supplement (e.g., 0.l mM), not with high (2 mM) (503, 1360, 1542). Pigment production has been used to assess the effect of histidine and tryptophan or purine nucleotide synthesis (1543). Reduced interallelic fertility (487, 771). Complementation maps (489, 501). Relation of mutagens to complementation patterns (490). Mutants with nonpolarized complementation patterns on the right side of the complementation map grow on minimal medium if supplied with CO2; other mutants do not respond to CO2 (491). Used in conjunction with ad-3A for studies of mutagenesis (see ad-3A). Rearrangement T(I®III)Y112M4i ad-3B, which has a breakpoint inseparable from ad-3B, was the first insertional translocation reported in fungi (485). Allele 7-017-0137 shows “fixed instability,” mutating to an unstable prototrophic allele (97). Alleles 2-17-126 and 12-21-28 and numerous others are supersuppressible amber mutants (772, 1461, 1860). Called complementation group B.
ad-4 : adenine-4
IIIR. Between met-8 (1%; 4%), com (0%; 5%) and col-16, leu-1 (1%; 3%) (426, 1591).
Cloned and partially sequenced: EMBL/GenBank AI416404; EST NC2G7.
Requires adenine. Cannot use hypoxanthine or inosine (1314). Growth on adenine (at least of allele 44206) is improved by the addition of histidine, and improved still more by the addition of histidine plus methionine (1314). Structural gene for adenylosuccinase (EC 4.3.2.2), which controls two reactions in adenine synthesis, namely, SAICAR to AICAR in the common part of the purine pathway and adenylosuccinate to adenosine 5’-P in the adenine-specific branch (249, 736, 2240) (Fig. 5). Accumulates a slight amount of purple pigment when adenine is limiting (1360). Used for the first demonstration of complementation between alleles in vivo (736) (simultaneous with independent demonstration in am) and in vitro (2239). Enzyme in revertants (2240). Used to study purine transport (1544). Alleles 44206 and 44415 are heat sensitive (34 vs 25ºC) (907, 1360), osmotic-remediable at 30ºC (1273); the enzyme synthesized at 30ºC by heat-sensitive strains has altered properties (1273). Originally called complementation group F.
ad-5 : adenine-5
IL. Between phe-1 and arg-1 (1%) (914, 1592).
Uses adenine or hypoxanthine (1360). Structural gene for a bifunctional enzyme encoding AICAR transformylase (EC 2.1.2.3) and inosine-5¢-monophosphate (IMP) cylohydrolase (EC 3.5.4.10). Accumulates AICAR (160, 1764) and SAICAR (160) (Fig. 5). By analogy with yeast, a putative isogene of ad-1. Some alleles are stimulated by histidine and may not grow on hypoxanthine unless histidine is present; others may be inhibited by histidine (295, 736). Produces some purple pigment, but less than ad-3A and ad-3B (997). Evidence of ref. (249), apparently enzymatic, suggests that some ad-5 mutants lack both the AICAR formyltransferase and IMP cyclohydrolase activities of the bifunctional polypeptide, but others lack only one or the other. Allele Y112M192, for example, is blocked only at the formyltransferase step (1763, 1764). Called complementation group J.
ad-6 : adenine-6
IVR. Between ilv-3 (9%) and pan-1 (1% or 2%), chol-1 (l%), cot-1 (2%; 6%) (1124, 1255).
Blocked in phosphoribosylformylglycinamidine synthase (FGAM synthase, EC 6.3.5.3) (249) (Fig. 5). Uses adenine or hypoxanthine (1360). Inhibited by caffeine in the presence of adenine (2279). Called complementation group I.
ad-7 : adenine-7
VR. Between cot-2 (4%) and T(EB4)R, ro-4 (4%), pab-2 (8%) (321, 322, 1578).
Cloned: Orbach-Sachs clones X22H08, X24H05
Uses adenine or hypoxanthine (1360). Lacks phosphoribosylpyrophosphate amidotransferase (EC 2.4.2.14), the first enzyme in de novo purine biosynthesis (996) (Fig. 5). Ascospores from homozygous ad-7 ´ ad-7 crosses are white (allele Y175M256). Allele P73B171(t) is temperature-sensitive.
ad-8 : adenine-8
VIL. Between het-8 (12%), ser-6 (l5%) and cpl-1 (7%; 11%), aro-6 (8%; 12%) (822, 963, 1425, 1582).
Requires adenine; cannot use hypoxanthine (997). Lacks adenylosuccinate synthase (IMP-aspartate ligase, EC 6.3.4.4) (964) (Fig. 5). Has little hypoxanthine uptake and low hypoxanthine phosphoribosyltransferase; both these effects are partly counteracted in the ad-1; ad-8 double mutant (1765). Low hypoxanthine phosphoribosyltransferase is also found in mep(3) and mep(l0). Used to study purine transport (1544, 1765). Fine-structure mapping and intralocus complementation (963-965). Called complementation group E.
ad-9 : adenine-9
IR. Between met-6 (9%; 16%), tre (7%) and T(T54M94)M, In(OY348)R, nit-1 (3%; 15%) (1578, 1592).
Cloned: pSV50 clone 6:3G (1819).
Uses adenine or hypoxanthine (997). Lacks phosphoribosylglycinamide formyltransferase (GAR transformylase, EC 2.1.2.2). Controls conversion of GAR to FGAR (249) (Fig. 5). Called complementation group D.
adg : adenine-arginine
Allelic with arg-11.
adh : adherent
VIIL. Between cyt-7 (9%) and nic-3 (4%; 11%). Linked to do (0/53), spco-4 (4%) (1582, 1592).
Morphology is abnormal. Conidia are not powdery and do not shake loose. Makes arthroconidia but few blastoconidia. Morphologically distinct from do and spco-4. Complements spco-4 (1582, 1592)
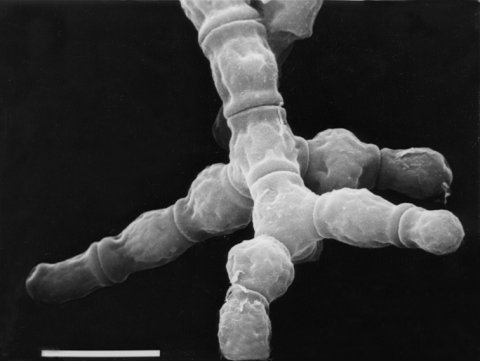
FIGURE 6 The mutant adh: adherent. Macroconidia of this type, called arthroconidia
("joint" conidia),
differ in mode of formation from blastoconidia ("bud" conidia), shown in Fig. 31B.
Bar length = 10 mm. Scanning EM photograph by M. L. Springer.
From ref. (1958),
with permission from Cold Spring Harbor Laboratory Press.
ads : adenine-sensitive
IV. Linked to col-4 (966).
Growth is completely inhibited at 35ºC by l0 mM adenine; high concentrations are inhibitory at 25ºC. Growth is poor on minimal medium at 35ºC, compared to 25ºC. Inhibition is not relieved by vitamins, amino acids, or by guanine, guanosine or guanylic acid; there is no growth response to guanosine in the absence of adenine (962, 966).
aga : arginase
VIIR. Between wc-1 (2%) and arg-10 (24%; 27%) (463, 1380). Near un-10 (1264).
Cloned and sequenced: Swissprot ARGI_NEUCR, EMBL/GenBank L20687, EMBL NCAGA, GenBank NEUAGA.
Structural gene for arginase (EC 3.5.3.1) (463, 1380) (Figs. 10-12). Unable to form ornithine from arginine; arginine is thus unable to satisfy the proline requirement of pro-3 in the pro-3; aga double mutant. The prototrophic single mutant develops a polyamine requirement in presence of arginine. This is due to feedback inhibition of ornithine biosynthesis by arginine, combined with a catabolic block in ornithine formation from arginine (458, 463). Two forms of arginase polypeptide (41- and 36-kDa) are produced (200). These differ at their N-termini and arise as a consequence of two different translation initiation codons being used in each of two differentially expressed mRNAs transcribed from alternative promoters (1264). The aga mutant has been used to study polyamine synthesis (466) and to obtain mutants defective in siderophore uptake (359); see sit. The triple mutant arg-12; ota; aga was used to look at deoxyhypusine and hypusine modification of a 21-kDa polypeptide (2261) that is the precursor of elongation factor eIF5A (see eif5A). Sideramine production is completely blocked in the absence of ornithine in the triple mutant arg-5; ota; aga, which is used to study iron transport (2227, 2229) and iron storage (1296, 1297).
age: aging of conidia
Used by K. D. Munkres for mutations showing reduced conidial longevity in the light. Not expressed in the dark or, with vitamin E or reduced glutathione, in the light. Deficient in enzymes that destroy free radicals and peroxides. Scored by plating efficiency after incubation of mature slant cultures at 30ºC, l00% relative humidity, in continuous white fluorescent light (24 Jm-2). Most age mutants can also be scored by a defect in conidiophore development on Vogel's sorbose-sucrose plates. The age mutants were initially selected as spontaneous variants from the fl of Oak Ridge wild types. Variants with increased conidial longevity also can be selected. Longer life span is correlated with slower growth (1394, 1395).
age-1 : aging of conidia-1
IR. Mutations designated age-1 were interpreted as being at a complex of recombinable sites distal to al-1, with individual sites being symbolized 1.1, 1.2, l.3, etc. Reversion or misscoring may possibly have led non-mutant progeny of intercrosses to be misinterpreted as crossovers (1394)
The short-lived phenotype of age-1 mutants is associated with abnormal morphology. Conidiophores are absent and conidia are formed on the agar surface. The various age-1 mutations are stated to be dominant to wild type and qualitatively indistinguishable from one another in longevity, morphology, and enzymatic defects. Mutations designated 1.3 are apparently alleles of so (1394).
age-2 : aging of conidia-2
VIR. Right of ws-1 (8%) (1394).
Conidiophores are absent, and conidia are formed on the agar surface. Dominant to wild type (1394).
age-3 : aging of conidia-3
An al-1 allele [also called ylo-4 (1394)].
agg : accelerated growth on galactose
Poorly defined genetically. Inheritance is not simple, with nuclear and perhaps cytoplasmic components (125). Best considered as designating a mutant strain, not a gene locus.
Differs from wild type in being able to use lactose as a sole carbon source in agitated cultures and in producing four times the wild-type amount of growth in standing cultures. Used to establish the existence of two b-galactosidases in Neurospora (127) and to study responses of the two enzymes to inducers (126). Called L5.
agr-1 : altered glucose repression-1
Unmapped.
Glucoamylase and other hydrolases that are normally repressed by glucose are synthesized and secreted constitutively (1913).
al : albino
Carotenoid synthesis is impaired in mutants designated albino Orange macroconidia and mycelia are characteristic of wild type strains in all of the conidiating Neurospora species. (Some Neurospora discreta and Neurospora intermedia strains are yellowish.) Carotenoids are also synthesized in microconidia (2120) and in perithecia and protoperithecia (1569), where their presence is normally masked but is revealed when the per mutation deletes black pigment. Different mutant albino alleles result in strains that are white, yellow, pink, or purple, that have only traces of color, or in which only peripheral conidia become pigmented (1570). An unstable constitutive variant has been described (1139). Carotenoids are also affected by genes designated ylo, by modifiers of intensity such as int and vvd, and by genes affecting induction by light (wc, ccb, lis, blr). Rapid development of carotenoids is induced by blue light (479, 837, 1378, 1760, 2295). Control by light is at the level of transcription initiation. For references and a review of photobiology, see ref. (1141). The blue light photoreceptor is still intact in the triple mutant al-1;al-2;al-3 (1760). Carotenoid production is maximized if dark-grown cultures are placed at ~5ºC immediately after exposure to inducing light (835). Albino mutants can be scored in submerged colonies if platings are grown at 25ºC in the dark and transferred to 5ºC under light (319, 933). Carotenoid synthesis is also developmentally regulated. Wild-type macroconidia slowly become pigmented in the dark, and conidia slowly become pigmented in the mutants wc-1 and wc-2, which are blind to photoinduction. For developmental regulation and photoregulation during conidiation, see refs. (1188) and (1189). For background and biochemical methodology, see ref. (1703). Albino mutants have been used to examine the role of carotenoids in photoprotection (1907, 2084). Epimutations of albino genes have been useful in studies of transgene-induced gene silencing (“quelling”) (390, 392, 1737). The most commonly recovered albino mutations are at al-1 and al-2 (700, 1845), which were once thought to be contiguous but are now known to be separated by other loci unrelated to carotenoid biosynthesis (1546, 1554). Cultures of osmotic mutants and of eas may become intensely orange; this can be ascribed to physical effects as cells adhere, rather than to changes in carotenoid production. Mutations that delete or modify the black pigmentation of perithecia and ascospores are not called albino but have been given other names, such as per, ws, and bs.
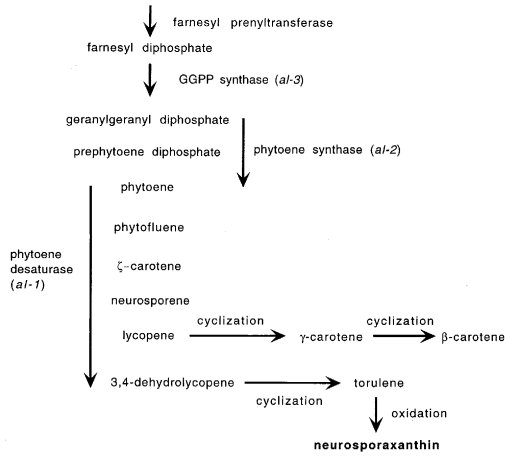
FIGURE 7 Carotenoid biosynthesis in Neurospora, showing positions of albino loci in
the pathway.
Original figure from T. J. Schmidhauser. A dual function is strongly suggested for al-2 by its
homology to a gene in Xanthophyllomyces that encodes a bifunctional protein involved both in
geranylgeranyl diphosphate and in cyclization of lycopene to -carotene and torulene
(2146a).
al-1 : albino-1
IR. Between hom (l%) and cnr (1%) (1554, 1578, 1580). On a common plasmid with hom (1816).
Cloned and sequenced: Swissprot CRTI_NEUCR, EMBL/GenBank M57465, PIR A35919, GenBank NEUAL1A, EMBL NCAL1A, pSV50 cosmid 3:11H, Orbach-Sachs clone G15C10.
Structural gene for phytoene dehydrogenase (phytoene desaturase) (EC 1.3.-.-) (1816), which catalyzes five steps in carotenoid synthesis (Fig. 7). Alleles differ widely in phenotype, ranging from white (e.g., allele 4637) and "aurescent" (pigment in peripheral conidia and conidiophores, allele 34508) to yellow mycelia and conidia (alleles ALS4, RES-25); see ref. (1570). Alleles ALSl4, RES-6, 34508, and RES-25 contain large amounts of phytoene (>99% of total neutral carotenoids) (748, 2018). Allele RWT-ylo accumulates ζ-carotene and smaller amounts of neurosporene, suggesting a leaky block between them (2084). A mutation called ylo-4 (also age-3) proved to be a pigmenting al-1 allele (1582). Early fine-structure mapping (933, 2020) needs to be reevaluated because the flanking marker arg-6 was positioned incorrectly (1554). For complementation tests, see refs. (933), (2018), and (2019). Used in studies of posttranscriptional gene silencing (quelling) (392, 1737). Translocation T(4637), which is inseparable from al-1, was the first albino mutant and one of the first chromosome rearrangements to be identified and studied in Neurospora (1307). For visualization of pachytene chromosomes and the synaptonemal complex in crosses heterozygous for T(4637), see refs. (1578). T(4637) was used to show that transcription of al-1 proceeds toward the centromere.
al-2 : albino-2
IR. Between cyh-1 (8%; 13%), T(STL76) and Tp(T54M94)R, arg-6 (1%), hom, al-1. Near os-5 (<l%) (1554, 1559, 1578, 1580, 1603, 1605).
Cloned and sequenced: Swissprot PSY_NEUCR, EMBL/GenBank L27652, PIR A53583, GenBank NEUAL2X, EMBL NCAL2X; Orbach-Sachs clones X14D06, X24C01, X25F09, G2G08.
Encodes phytoene synthase (EC 2.5.1.-) (1817), a particulate enzyme (838) (Fig. 7). Regulated developmentally and by light. Phenotypes of different alleles range from white and pale rose-white for l5300 and Y254M165 (2020) to purple for MN58a (319). For complementation, see refs. (933) and (2019). Fine-structure mapping (933, 2020) needs to be reevaluated because the flanking marker arg-6 was incorrectly located (1554).
al-3 : albino-3
VR. Between pho-2 and inl (l%; 1T/18 asci) (747, 1447, 1582, 2185).
Cloned and sequenced: Swissprot GGPP_NEUCR, EMBL/GenBank U20940, X53979, PIR S15662; pSV50 clones 23:1A, 23:1A, Orbach-Sachs clone X17:A07 (1451).
Structural gene for the trifunctional enzyme encoding geranylgeranyl pyrophosphate synthase (EC 2.5.1.1), geranyltransferase (EC 2.5.1.10) and farnesyltransferase (EC 2.5.1.29) (2161) (Fig. 7). Encodes two overlapping transcripts, AL-3(m), expressed in mycelia following light induction, and AL-3(c), induced developmentally and by light, and subject to circadian control (58). AL-3(c) is photoregulated only early in conidiation, unlike transcripts of other albino genes (1188). Allele Y234M470 (al-3ros), formerly called rosy (105), becomes partially pigmented but is readily distinguished from wild type. ylo-1 can be scored in al-3ros (Y234M470) (1582). Other alleles (e.g., RP100) (2185) are white with a trace of pink pigment. Null mutations are probably lethal [N. Romano unpublished, cited in ref. (1737)], as might be expected if al-3+ function is required for viability. Placement of al-3 early in the biosynthetic pathway is consistent with its having another function that is essential. Compounds affecting protein kinase C activity alter the response of al-3 to light, implicating protein kinase C in al-3 regulation (59). Among progeny of strains duplicated for al-3+, the only surviving mutants are those that have undergone mild RIP (95).
alc-1 : allantoicase-l
IIR. Linked to pe (10%), probably on the opposite side of pe from xdh-1 (24%) (1712). Linked very close to arg-12 by RFLP (1277).
Cloned and sequenced: Swissprot ALC_NEUCR, EMBL/GenBank J02927, PIR A35829, GenBank NEUALCA, EMBL NCALCA.
Defective in purine catabolism. Unable to use allantoic acid as sole nitrogen source. Structural gene for allantoicase (EC 3.5.3.4) (1712) (Fig. 8); for regulation, see Fig. 50. alc-1 expression requires induction, and this requires a functional nit-2 product, with binding of NIT2 to two GATA elements in the alc-1 promoter (1151).
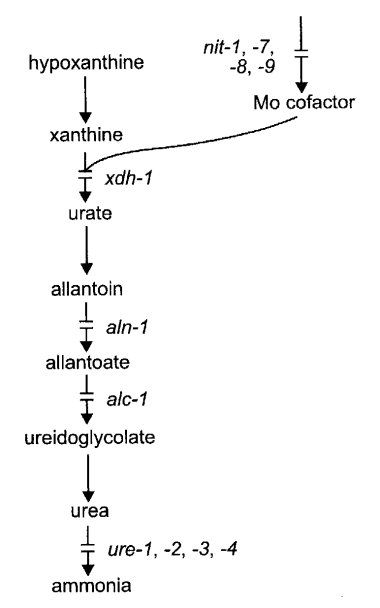
FIGURE 8 The purine catabolic pathway, showing sites of action of the xdh, aln, alc,
and ure genes
(
862,
1087 ,
1712).
The molybdenum cofactor is essential to both xanthine dehydrogenase and nitrate reductase
(2102).
From ref. (1596 ),
with permission from the American Society for Microbiology.
alcoy (acronym for al-1; cot-1; ylo-1)
Genotype: T(IR;IIR)4637 al-l; T(IVR;VR)R2355, cot-1; T(IIIR;VI)l, ylo-1 (1592).
A linkage tester strain containing three unlinked reciprocal translocations, each tagged with a visibly scorable marker, marking linkage groups I-VI (Fig. 9). Linkage of a gene to al-1, cot-1, or ylo-1 in a cross to alcoy allows assignment to linkage group by a single follow-up cross. A majority of new point mutants are linked to one of the alcoy markers (1592). An improved version, alcoy; csp-2, carries the VII marker csp-2 in addition to the three original markers (1572, 1584). alcoy strains been used cytologically in studies of the synaptonemal complex and recombination nodules (740, 1231).
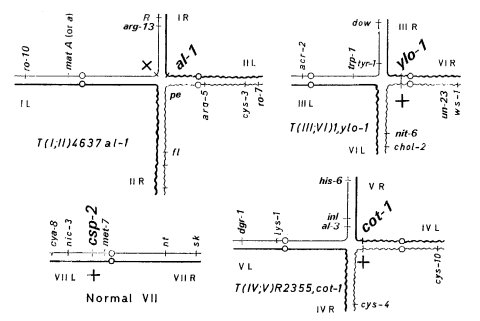
FIGURE 9 Linkage relations in a cross of the linkage tester alcoy; csp-2 X Normal sequence. Chromosomes are shown paired as in meiotic prophase I. The alcoy; csp-2 chromosomes are drawn as heavy lines, and those from the standard-sequence parent are shown as thin lines. Segments from odd-numbered linkage groups are shown as straight lines and even-numbered as wavy lines. The only mutant markers present in the alcoy parent are al-1, cot-1, ylo-1, and csp-2. These are shown in large letters. al-1 is inseparable from the T(I; II)4637 breakpoint in linkage group I, cot-1 is a few units proximal from the T(IV; V)R2355 breakpoint in IV, and ylo-1 is a few units from the T(III; VI)1 breakpoint in V (arbitrarily shown as centromere proximal). Also shown are the locations of markers present in the standard-sequence follow-up testers, a few key markers for orientation, and some useful distal markers that are far removed from the breakpoints. The latter are used only when no linkage to the alcoy markers is detected. Interval lengths and relative distances are not to scale, but gene order is as shown. Figure from D. D. Perkins.
aln-1 : allantoinase-l
VII (1712).
Defective in purine catabolism. Unable to use allantoin or any purine intermediate prior to it as the sole nitrogen source. Lacks allantoinase (EC 3.5.2.5) (1712) (Fig. 8); for regulation, see Fig. 50.
Alpha-tubulin
See tba.
alx-1 : alternate oxidase-1
Used for a double-mutant strain called ANT-1.
alx-2 : alternate oxidase-2
Unmapped.
Lacks inducible cyanide-insensitive respiration. Cannot grow on antimycin A. Complements the double-mutant strain called alx-1 or ANT-1 (591).
am : amination-deficient
VR. Between sp (4%; 8%, 450 kb), ure-2 (2%) and gul-1 (<1%), ace-5 (<1%), ure-1 (1%; 150kb), his-1 (3%) (217, 219, 220, 256, 1086, 1124, 1941).
Cloned and sequenced: Swissprot DHE4_NEUCR, EMBL/GenBank K01409, PIR A00381, DENCEN, GenBank NEUAMG, EMBL NCAMG, EST NM2B4; pSV50 clones 12:2A, 15:1B, 17:2A, 32:3H, Orbach-Sachs clones X1C05, X5F02, X6B01, G7G02, G9A10, G9D12.
Structural gene for a NADP-specific glutamate dehydrogenase (EC 1.4.1.4) (639) (Fig. 49). Requires a source of a-amino nitrogen for growth; alanine is a good supplement (1940). Readily scorable at 25ºC; leaky at 34ºC (100). Leaky growth and adaptation on minimal medium are prevented by 0.02 M glycine (1536, 1537) or by en(am)-1, en(am)-2, or nit-2. The am mutants show abnormal regulation of NADH-glutamate dehydrogenase and are synergistic with nit-2 in this effect (442). Some am alleles (e.g., RU1) suppress the pyrimidine requirement caused by pyr-3 (CPS-ACT+) mutations (2212). Used for the first demonstration of complementation between alleles in vivo (646) (simultaneous with independent demonstration in ad-4). In vitro complementation (640). Used for studies of the complementation mechanism (386, 387, 2191) and for fine-structure mapping (634, 641). Used to study the mechanism of recombination (218, 221), including the control of intralocus recombination by rec-3 (1939-1941), the relation of gene conversion and crossing over (219), and the orientation of the fine-structure intragenic map relative to flanking markers (217). Used to study the mechanism of RIP (636) and to generate new alleles by RIP (643). Instrumental in discovery of the Tad retrotransposon (1065). Used to study colinearity of gene and gene product, internal suppressors (226, 635, 888), and the action of amber supersuppressors (1859, 1860). Used as a site for targeting homologous integration of transforming DNA (1343), for transposon targeting (277), and for the identifying upstream regulatory regions (371, 670, 671). The functional defects in several mutant enzymes with single amino acid replacements have been defined. The product of am1 fails to bind NADPH (2191); those of am2, am3, am19, am130, and am131 are stabilized in an inactive conformational form (66, 386, 639, 1063, 2023) and all are complementable by am1. Allele am14 is osmotically reparable and thought to have unstable quaternary structure (635). Used in a study showing glutamine to have a role as co-repressor of uricase synthesis (2184). Used to study nitrogen assimilation and metabolism (939) and nitrogen metabolite repression (354, 564). Efficient procedure for selecting new am mutants (1058). am mutants suppress the p-phenylalanine resistance of mtr (1058) and fpr-6 (1335). Spectrum of UV- and nitrous acid-induced mutants (1066). Used to study the effect of mutants on intron processing (1054) and the effect of mutations to rare codons (those with A in the third position) in reducing translation efficiency (1053). Allele am17 has a chain-terminating amber codon at residue 313 of GDH, based on amino acid replacements in revertants and ssu1 (1861). Allele 6 is a frameshift with insertion in Ser5 codon (1912). Allele 126 is highly unstable (1062). Allele 132 is a deletion (2245). In(UK2-y), T(UK9-18), T(mpr13-1), and T(mpr15-2) are inseparable from am (1578).
amdS : acetamide utilization
Introduced from Aspergillus nidulans.
Encodes an acetamidase that confers the ability to use acetamide as the sole nitrogen source. This Aspergillus gene has been used in preference to the Neurospora gene Bml as a dominant selectable marker for transformation (2257). Transformants containing a single copy of amdS are not inactivated by RIP and, therefore, can be recovered from crosses. In contrast, when the essential Neurospora gene Bml is used as a selectable marker, transformed genes cannot be readily recovered from crosses because the functional Bml+ allele is inactivated by RIP. See also ble.
amr : ammonium regulation
Allelic with nit-2.
amt : aminotransferase
VR. Between crp-4 (8T/18 asci) and Tel-VR (2T/18 asci) (1452).
Cloned and sequenced: cDNA clones NM8H12, NP2A7.
Encodes putative amidinotransferase (EC 2.1.4.1) (875a, 1408).
amy-1 : amylase-1
Allelic with sor-4.
amy(SF26) : amylase(SF26)
Allelic with exo-1.
amyc : amycelial
IL Between ad-5 and Cen-I (914).
Recessive. On sucrose media, amyc forms dotlike granular colonies of irregular budding vesicular elements. On permissive media, made either with acetate (plus a-ketoglutarate, succinate, malate, or certain amino acids) or with amino acids as C and N sources, it forms hyphae and macroconidia but apparently is still colonial (532, 2117). cAMP induces conidiation even on sucrose (532). Conidia show multipolar germination (2118). Photographs (1510, 2117). For low oxygen consumption and depressed amino acid pools, see (532), abnormal mitochondria (1509), surface glycopeptide (532), wall composition (403), recovery of antigenic arc representing the isozyme of malate dehydrogenase associated with conidiation (1539). Ultrastructure (1509, 2117, 2119). Used extensively in a balanced heterokaryon to evaluate nuclear distribution (71) and to detect lethal recessive mutation (70), anticipating the method of Stadler (1980). Techniques are described in ref. (72).
an : anaerobic
Unmapped.
Reported to be facultatively anaerobic, showing weak growth on enriched medium in the absence of oxygen. Prototrophic and indistinguishable from wild type under aerobic conditions. Not glucose-repressed. Anaerobic cultures aconidial, with reduced cytochrome oxidase and malate dehydrogenase activities, mitochondrial changes, and production of ethanol. Obtained by filtration enrichment with recycling. The symbol An+ was used to specify the mutant phenotype, An- the wild phenotype (922, 923). Strains deposited in FGSC as anaerobic mutants have been tested unsuccessfully for their ability to grow under strictly anaerobic conditions (162).
ANT-1 : antimycinsensitive
Not a locus designation. The symbol ANT-1 was used (595) to designate the double-mutant strain azs; has, which was also called alx-l. See azs and has.
anx14 : annexin 14
Unmapped.
Cloned and sequenced: GenBank AF036871.
Encodes annexin XIV (225). Related to annexins in Dictyostelium and in animals. (No annexins are found in Saccharomyces cerevisiae.)
aod-1 : alternate oxidase deficient-l
IV. Linked to Cen-IV (0T/17 asci) (1447). Left of trp-4 (23%) (164, 2033).
Cloned and sequenced: Swissprot AOX_NEUCR, EMBL/GenBank L46869, PIR S65752, GenBank NEUALTO; pSV50 clone 23:7F.
Structural gene for the alternate terminal oxidase that is used instead of cytochrome c oxidase when cytochrome-mediated, cyanide-sensitive respiration is inhibited (1192). AOD-1 is identified immunologically as Mr 36,500 and 37,000 polypeptide species present in high concentrations under inducing conditions (1135). Alternate oxidase activity can be stimulated by some, but not all, nucleotides (1344, 1524, 2142). Strains carrying aod-1 plus either [mi-1] or [mi-3] are viable (165, 2033). Called aod-B (165). Three recessive mutant alleles, originally called aod-1, aod-2, and aod-3, later were called aod-1.1, aod-1.2, and aod-1.3 (2033).
aod-2 : alternate oxidase-2
II. Linked to arg-5 (7%), thr-3 (16%), trp-3 (36%) (164, 2033).
The alternate oxidase system fails to be induced when cytochrome-mediated, cyanide-sensitive respiration is inhibited (164). The aod-2 defect appears to eliminate the increase in the level of aod-1 transcript observed under these conditions (1192).
Aph-1 : Aphidicolin resistance-1
Unmapped. Mutant alleles show Mendelian segregation (1354).
Resistant to aphidicolin; an inhibitor of DNA polymerase a at 200-800 mg/ml. Resistance is dominant. With mutant strain E2-4-1, enzyme activity in vitro is resistant to aphidicolin inhibition. With mutant C-3, the pH optimum for enzyme activity is changed. Mutations were obtained by plating nitrosoguanidine-treated slime in medium containing 20 mg/ml aphidicolin. Stocks of AphR; slime have been kept as heterokaryons on minimal agar medium (1354).
apu : accumulation of purines
Unmapped. Not linked to ad-7 (VR) or mat, aza-1 (IL).
Excretes purines. Obtained among prototrophic revertants of ad-7 (which blocks the first step in de novo purine synthesis). Secretion was assayed by cross-feeding on plates seeded with ad-3A conidia. Purine secretion by apu occurs later than with aza-1 allele 67-12 (4 days vs 36 hr, 25ºC) and colony size is larger (996).
ar-2 : (mating type) A/a related
Allelic with eat-2.
arg: arginine
For details of the arginine biosynthetic pathway, see Fig. 10. The most comprehensive references on arginine are (457) and (458). Arginine mutants have been used extensively for studies of compartmentation (436, 471, 475) and for studies of control of flux through the arginine pathway (468) (also see ota entry). Crossing is inhibited by high arginine levels; 0.l or 0.2 mg/ml in crossing medium is satisfactory. Lysine and arginine show competitive inhibition, and all arginine auxotrophs are inhibited by lysine. Crosses or strains involving both requirements usually can be handled by adjusting the ratio; 0.8 mg/ml L-arginine-HCl:l.6 mg/ml L-lysine-HCl is recommended for crosses (1970). Leaky arginine mutants (e.g., arg-2, arg-3, arg-13) are less leaky on nitrate medium (457) or canavanine plus lysine (1716). Leakiness of germinating ascospores of arg-1 and arg-3 is prevented by 0.05 mg lysine/ml with no canavanine (1476). Lysine resistance is conferred on arg-1 by a probable transport mutant lysR . Some arg genes were originally called cit or orn. For degradative or related steps in arginine metabolism, see aga, ota, spe, and ure.
Regulation: Arginine biosynthesis and catabolism are controlled in a major way by compartmentation (455, 458) (Figs. 11, 12). Acetyl glutamate synthase and acetyl glutamate kinase are feedback-regulated by arginine (458). With one exception, the enzymes of arginine biosynthesis are not repressed below levels seen in minimal medium when arginine is added to cultures. The exception is carbamoyl phosphate synthetase A (CPS-A), the arg-2-encoded small subunit of which is repressed 5-fold. Upon limiting cultures for arginine, all biosynthetic enzymes except one increase concomitantly by about 3- to 5-fold; the CPS-A small subunit increases by as much as 20-fold (458, 677). These "derepressions" can also be brought about by starvation for other amino acids, such as tryptophan, lysine, and histidine; they require the normal product of the cpc-1 locus (458, 1768). See cpc-1. The catabolic enzymes, arginase and ornithine aminotransferase, are present without induction and are elevated by only 2- to 5-fold in response to nitrogen limitation or the addition of arginine (but not ornithine) to the medium. These enzymes are not affected by mutations at the nit-2 and/or cpc-l loci.

FIGURE 10 The biosynthetic pathways for arginine, proline, polyamines, and associated
intermediary metabolism,
showing sites of gene action in biosynthesis and arginine catabolism
(457 ,
465 ,
862 ,
1087 ,
1310 ,
1379 ,
1380 ,
1967 ,
2165 ,
2291 ).
Carbamoyl phosphate for pyrimidine synthesis is made as a separate pool by a distinct enzyme
(see Fig. 53).
Interchange between the two pools occurs only in certain mutant combinations. From ref.
(1596 ),
with permission from the American Society for Microbiology. For details of the polyamine
pathway, see Fig. 62.
arg-1 : arginine-1
IL. Between ad-5 (1%) and eth-1 (<1%) (1462, 1592).
Cloned: pSV50 clones 9:11F, 30:6G (1818).
Uses arginine, but not its precursors (1968). Lacks arginosuccinate synthetase (EC 6.3.4.5) (1463) (Figs. 10 and 11). Interallelic crosses produce perithecia, but most ascospores are white and inviable (1462). Leaky arg-l mutants are frequent among mutants selected as citrulline-resistant variants of arg-12S; pyr-3; most of these show interallelic complementation, and many are transport-deficient (2089). arg-l does not grow well on some complex complete media unless extra arginine is added.
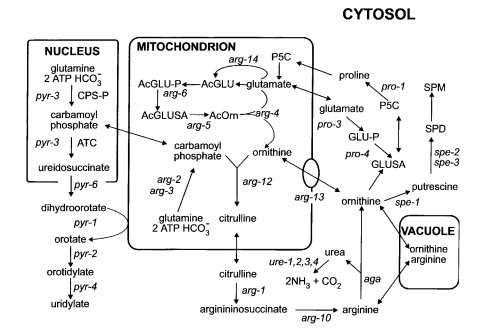
FIGURE 11A schematic view of the organization of arginine, pyrimidine, proline, and
polyamine metabolism in
N. crassa. Boxes enclose cellular organelles to delineate the reactions carried out within
different
cellular compartments. The names of genes are shown next to the metabolic steps executed by
their products.
The product of arg-13 is positioned on the mitochondrial membrane, where it is predicted to act
as a carrier.
The genes pro-3 and pro-4 originally were called arg-8 and arg-9. Abbreviations: ATC,
aspartate
carbamolytransferase; CPS-P, pyrimidine-specific carbamoyl-P synthetase; Ac, acetyl; GLU,
glutamate; GLU-P,
glutamyl phosphate; GLUSA, glutamate semialdehyde; ORN, ornithine; P5C,
pyrroline-5-carboxylate; SPD, spermidine;
SPM, spermine. Adapted from refs.
(1213 ) and
(458 ),
with permission from the Genetics Society of America and the American Society for
Microbiology, respectively.
arg-2 : arginine-2
IVR. Between col-4 (l%; 2%) and T(S4342)L, pyr-3 (l%; 3%) (196, 1369, 1372, 1578, 1580, 1596, 1716, 1928).
Cloned and sequenced: Swissprot CARA_NEUCR, Y3KD_NEUCR, EMBL/Genbank J05512, EMBL NCPSAS, GenBank NEUPSAS; pSV50 clones 6:7C, 11:7E, 20:5B, 24:8D.
Uses citrulline or arginine (1968). Structural gene for the small subunit of arginine-specific carbamoyl phosphate synthetase (EC 6.3.5.5), a two-subunit enzyme (471). The small subunit enables the enzyme to use glutamine as a nitrogen donor (458, 468). (The large subunit is specified by arg-3; Figs. 10 and 11.) For regulation and compartmentation, see ref. (458). Arginine-specific negative translational regulation requires an evolutionarily conserved upstream open reading frame in the mRNA transcript that causes ribosome stalling (2187). The arginine-citrulline requirement can be suppressed by pyr-3 (CPS+ACT-) mutations (455, 1716); see pyr-3. Leakiness is prevented by canavanine; lysine overcomes the side effects of canavanine (1716). At least some alleles can grow on minimal medium in 30% CO2 (230). Leakiness is decreased if CO2 is removed or if uridine is added (1721). Translocation T(MEP24) is inseparable from arg-2 (451).
arg-3 : arginine-3
IL. Between eth-l (1%) and csp-1 (1%), T(39311)R (1592, 1882).
Cloned and partially sequenced: GenBank AI392110, EST NC3A12, pSV50 clones 9:12E, 10:6C, 13:6A, 15:5E, 26:11B, 28:6G, 28:12D; Orbach-Sachs clones X1F03, X8:D04, X9C06, X10H06, X17B01, X17D07, X22H02, G1A11, G3E09, G4G05, G6F06, G9C04, G13C04.
Uses citrulline or arginine (1968). Structural gene for the large component of arginine-specific carbamoyl phosphate synthetase (EC 6.3.5.5), a two-subunit enzyme (458). This component can form carbamoyl phosphate in vitro, using ammonia as nitrogen donor (458). (The small component is specified by arg-2; Figs. 10 and 11). For regulation and compartmentation, see ref. (458). The arginine-citrulline requirement can be suppressed by pyr-3 (CPS+ACT-) (455, 1312); see pyr-3. Allele 30300 can grow on minimal medium in 40% CO2 (230, 363). Translocation MEP35 is inseparable from arg-3.
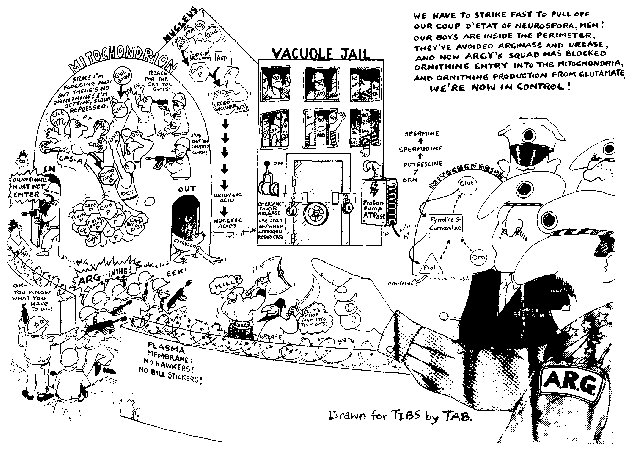
FIGURE 12 An alternative view of compartmentation involving arginine and related metabolites. Reprinted from ref. (475 ) [Davis, R. H., and R. L. Weiss (1988). Novel mechanisms controlling arginine metabolism in Neurospora. Trends Biochem. Sci. 13: 101-104.] Copyright 1988, with permission from Elsevier Science.
arg-4 : arginine-4
VR. Between sp (1%; 11%) and inl (2%; 4%) (1585, 1895).
Uses ornithine, citrulline, or arginine (1968). Lacks acetylornithine-glutamate transacetylase (EC 2.3.1.35) (acetylornithine acetyltransferase) (514, 2166) (Figs. 10 and 11). Weakly suppresses CPS-ATC+ pyr-3 mutants (1313). Alleles 21502 and 34105 (later called arg-4 and arg-7) were originally thought to be genetically distinct because they complemented (1963), but an intercross produced no recombinants (1313) and both lack the same enzyme (514, 2166).
arg-5 : arginine-5
IIR. Between bal (1%; 9%), T(ALS176) and aro-3, pe (6%; 18%) (800, 1546, 1548, 1578).
Structural gene for acetylornithine transaminase (EC 2.6.1.11) (1379) (Figs. 10 and 11). Uses ornithine, citrulline, or arginine (1968). Sideramine production is completely blocked in the absence of ornithine in the triple-mutant strain ota; arg-5; aga, which has been used to study iron transport (2227, 2229) and iron storage (1296, 1297). Called orn-1.
arg-6 : arginine-6
IR. Between al-2 (1% or 2%), T(T54M94)R and hom (1%), al-1 (l%;4%) (1554, 1578, 1580).
Cloned and sequenced: Swissprot AR56_NEUCR, EMBL/GenBank L27746, PIR A53429, EMBL NCPPOA, GenBank NEUPPOA, EST NC1G8; Orbach-Sachs clones X3A12, X13G12, X17D10.
Uses ornithine, citrulline, or arginine (1968). A bifunctional gene, specifying arginine-sensitive acetylglutamate kinase (EC 2.7.2.8) (457) and N-acetylglutamyl phosphate reductase (EC 1.2.1.38) (Figs. 10 and 11). The polyprotein precursor has an N-terminal mitochondrial targeting sequence followed by the kinase domain, a connecting region, and the reductase domain. Both the N-terminal-targeting sequence and the connecting region are cleaved by the mitochondrial-processing peptidase and
-processing-enhancing protein to yield two mature enzymes in the mitochondrion. Processing of the polyprotein precursor has been reconstituted in vitro (1535). L-Methionine may inhibit (1546). Can be selected as a suppressor of pro-3 by virtue of the feedback insensitivity of ornithine synthesis (2204). For supersuppressible amber alleles, see ref. (474). Called orn-2.
arg-7 : arginine-7
Allelic with arg-4. Also called orn-3.
arg-8 : arginine-8
Changed to pro-3.
arg-9 : arginine-9
Changed to pro-4.
arg-10 : arginine-10
VIIR. Between dr (12%), T(5936), arg-11 (1% or 2%) and nt (1%; 12%) (1462, 1548, 1578).
Uses arginine, but not its precursors (1462). Lacks argininosuccinate lyase (EC 4.3.2.1) (644) (Fig. 10). Accumulates arginosuccinate on limiting arginine (644). Viable ascospores from interallelic crosses are rare but the viable ones are often arg+, whereas most arg- ascospores remain colorless and inviable (1462). All arg-10 alleles tested showed spasmodic growth in growth tubes at low arginine concentrations (738). arg-10 does not grow well on some complete media unless extra arginine is added.
arg-11 : arginine-11
VIIR. Between dr, T(5936) and arg-10 (1% or 2%) (1548, 1578).
Requires arginine or citrulline, plus low levels of a purine and a pyrimidine (556, 1465, 1964). Inhibited by guanidine, sarcosine, and serocyamine (1963). Complements arg-l0 fully in heterokaryons (1465). The relation of this mutant to arginine biosynthesis or metabolism is obscure. Growth requirements vary markedly with CO2 concentration and inoculum size. At 0% CO2 or with small inocula, the requirement for all three supplements is absolute; with increasing CO2 concentration or inoculum size, pyrimidine and then purine can be omitted. At 30% CO2, all three supplements can be omitted (230, 364, 1465). Growth rate and morphology are highly variable among progeny from arg-11 ´ wild type (1465). Grows spasmodically in growth tubes (738). Allele 44601 was formerly called un(44601) (907) and adg (556).
arg-12 : arginine-12
IIR. Between pe (1%;5%) and T(NM177)R, aro-1 (<1%) (731, 1580, 2242).
Cloned: pSV50 clone 16:9H (699).
Structural gene for ornithine carbamoyl transferase (OCT; EC 2.1.3.3) (452, 473, 2242). Uses citrulline or arginine (Figs. 10 and 11). The leaky allele arg-12S, discovered as a suppressor of a pyr-3 mutant and initially called s (908), reduces OCT activity over 98% without imposing any arginine requirement. arg-12S suppresses the pyrimidine requirement of pyr-3 alleles that lack only pyrimidine-specific carbamoyl phosphate synthetase (CPS). This is because arg-12S accumulates arginine-specific carbamoyl phosphate, which can then be used for pyrimidine synthesis (455) (see pyr-3). Nonleaky arg-12 alleles cannot cause such suppression because the exogenous arginine that is required for growth results in repression of the arginine-specific CPS (455). Arginine mutants at all other arginine biosynthetic loci can be obtained efficiently as tight double mutants, using arg-12S as starting material (457). The double mutants pro-4; arg-12S and pro-3; arg-12S are prototrophic, due to diversion of ornithine to the proline pathway. The double mutant arg-5; arg-12S cannot use exogenous ornithine (453, 465).
arg-13 : arginine-13
IR. Between os-1 (1%) and so (2%; 12%) (1592).
Cloned and sequenced: Swissprot AR13_NEUCR, EMBL/Genbank L36378, EMBL NCARG13A, GenBank NEUARG13A; pSV50 clones 7:12B, 21:10D, 24:10H.
Structural gene for a nuclear-encoded protein that transports ornithine across the mitochondrial inner membrane (1213) (Fig. 11). Homologous with the human mitochondrial ornithine transporter responsible for hyperornithinaemia-hyperammonaemia-homocitrullinuria syndrome (274). Responds well to arginine or citrulline, but poorly to ornithine (457, 1313). Acts as a suppressor of the pyrimidine requirement of CPS-ACT+ mutations of pyr-3 (1313). Leaky on minimal medium; scoring is cleared by the addition of lysine. Interallelic crosses are sterile. Called arg(RU3).
arg-14 : arginine-14
IVR. Between T(S4342)L, arg-2 (1%) and T(NM152)L, pyr-3 (1%) (457).
Cloned and sequenced: EMBL/Genbank L35484, EMBL NCARG1A, GenBank NEUARG1A; pSV50 clones 7:12B, 21:10D, 24:10H.
Structural gene for N-acetylglutamate synthase (EC 1.4.1.13). Uses arginine, citrulline or ornithine (Fig. 11). Unlike the genes specifying other arginine biosynthetic enzymes, expression of arg-14 appears relatively constant over the course of the asexual life cycle (2287). Point mutants were selected as tight double mutants using arg-12S (457). Allele S1229 is inseparable from translocation T(S1229) (111, 112, 1580).
arg(CD-15)
Changed to cpc-1.
arg(CD-55)
Changed to cpc-1.
arg(RU1)
Allelic with am.
argR : arginine resistant
Allelic with pmb.
aro : aromatic
Used for genes concerned with biosynthesis of aromatic amino acids and p-aminobenzoic acid (PABA). See Fig. 13 for pathway and sites of gene action. Excepting aro-6, -7, -8, the genes designated aro are auxotrophs able to grow on a mixture of PABA, tyrosine, tryptophan, and phenylalanine. The first step in the pathway is catalyzed by three isozymes subject to feedback by different end products of the branched pathway and specified by different widely separated genes (aro-6, -7, -8). The second, third, fourth, fifth, and sixth steps are specified by a cluster gene that produces a single transcript (728, 2205), which produces a pentafunctional polypeptide (389, 2205). (The cluster gene is here called aro-1. Domains specifying the five functions were named aro-1, aro-2, aro-4, aro-5, and aro-9 before it was realized that they were not separate genes.) The final step prior to branching is specified by a unifunctional gene (aro-3), which is at a separate chromosomal locus from the aro cluster gene although linked to it at a distance. The third and fourth steps are paralleled by similar reactions in the quinate catabolic pathway (Figs. 13 and 54). Thus, the aro-9 enzyme ARO-9 can be replaced by QA-2, and under appropriate conditions, ARO-1 can be replaced by the QA-3. Supplement levels: 40-80 mg/l each tyrosine, tryptophan, and phenylalanine; 0.25mg/l PABA (344, 800). Also called arom.
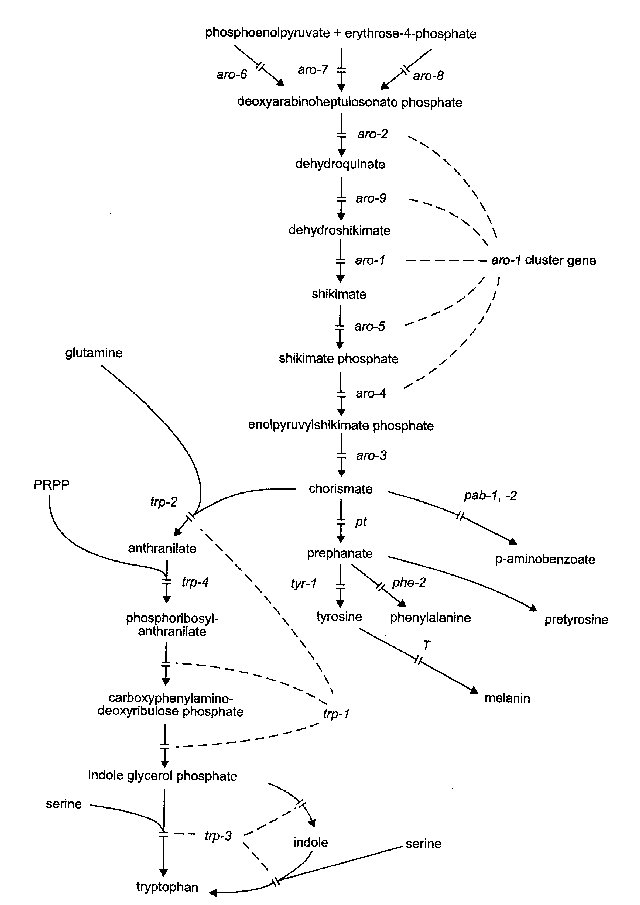
FIGURE 13 The biosynthetic pathways of the aromatic amino acids, showing sites of
action of the aro, trp, pt,
phe, tyr, and T genes (
193 ,
305 ,
396 ,
523 ,
601 ,
728 ,
822 ,
896 ,
999 ,
1038 ,
2227 ,
2264 ).
The conversion of chorismate to p-aminobenzoate has not been demonstrated in Neurospora.
In the conversion of tyrosine to melanin, the later steps are nonenzymatic. Products of the trp-1
and trp-2 genes form an enzyme aggregate with three properties: a given trp-1 mutant may block
one or more of the reactions; aro-9+ activity (biosynthetic dehydroquinase) can be replaced by
the product
of qa-2+, the equivalent gene in the catabolic pathway
(1726 );
and pretyrosine accumulates when other pathways are blocked (see phe-2). PRPP:
5-phosphoribosyl
pyrophosphate. From
ref. (1596 ),
with permission from the American Society for Microbiology.
aro-1 : aromatic-1 cluster gene
IIR. Between arg-12 (<1%), T(NM177)R and ff-1 (4%; 6%) (1102, 1578, 1580, 2044). Intracluster map shows the order of domains to be 1, 9, 5, 4, 2 (728, 1726).
Cloned (315); pSV50 clone 31:9G.
Structural gene for the aromatic biosynthetic pathway leading to tryptophan, tyrosine, phenylalanine, and PABA (Fig. 13). A multifunctional "cluster gene" (705) specifying five enzymes (389, 705, 731, 2205). Clustering of functions was discovered by (800). Reviews are in refs. (728) and (1325). The order of regions that specify the five functions is still conveniently represented by the symbols established when it was thought that five separate genes were involved: (arg-12) ... aro-1 aro-9 aro-5 aro-4 aro-2 ... (ace-1). These symbols actually represent five domains of the pentafunctional polypeptide, specifying products that catalyze five different steps in the pathway. The native enzyme is a dimer of the pentafunctional chains (389, 705, 1235). We now designate the entire cluster gene as a single locus, aro-1. Mutants exist that block individual steps; also polar mutants exist, symbolized aro(p), that eliminate more than one function. There are some discrepancies between genetic mapping and mapping by polarity [reviewed in ref. (1325)]. Genetic data indicate that transcription begins at the aro-2 end (303, 728). aro-1, -2, and -9 can use shikimate (0.3 mg/ml) as an alternative to the mixture of four aromatics shown for aro-1 by (2057). Mutants that block different individual steps complement with each other in heterokaryons (731). Complementation between alleles that block the same single step has been detected only for aro-2 and aro-1 (297, 731). Polar mutants are divided into six classes (A-F) based chiefly on their complementation behavior (731); types D-F are semicolonial with yellowish-orange conidia (294). Single-function aro-9 mutants were obtained by selecting in qa-1, which is noninducible for catabolic dehydroquinase activity (1726). Translocation T(C161)aro (called arom-2) is inseparable from the aro-1 cluster and lacks several activities (800). Noncomplementing alleles M26, M1039, M1065, M1108, M1162, M1172, and Y306M54 (abbreviated M54) are suppressible by amber suppressors (ssu- "supersuppressors") (294, 303, 1862). Details regarding individual components are given in the following paragraphs.
aro-1 domain: Specifies dehydroshikimate reductase (EC 2.5.1.19) (731, 800). Accumulates dehydroshikimate, which induces dehydroshikimate dehydrase in the catabolic pathway (800). Suppressed by qa-4, which lacks dehydroshikimate dehydrase; this allows induction of the catabolic enzyme quinate (shikimate) dehydrogenase, which substitutes for the biosynthetic enzyme dehydroshikimate reductase (305). A lag in the growth of aro-1 on shikimate occurs with sucrose or glucose as the carbon source. This is overcome by substituting 1% glutamate for the sugar (942).
aro-2 domain: Specifies dehydroquinate synthetase (EC 4.6.1.3) (731). aro-2 point mutants should not be confused with strain C161, called arom-2 (800), which lacks several activities in the aro-1 cluster, including aro-2 function (800), and is inseparable from translocation T(C161) (1580).
aro-4 domain: Specifies 3-enolpyruvate shikimic acid-5-phosphate synthetase (EC 4.2.1.10) (731).
aro-5 domain: Specifies shikimate kinase (EC1.1.1.25) (731).
aro-9 domain: Cloned. Active site sequence PIR S14749; pSV50 clone 31:9G. Encodes biosynthetic dehydroquinase (EC 2.7.1.71) (1726). Requires shikimic acid or a mixture of four aromatic products when a qa mutant is present that eliminates catabolic dehydroquinase. aro-9; qa+ grows on minimal medium without supplement. Single-function aro-9 mutants were obtained first by selecting in qa-l, a regulatory mutant that lacks catabolic dehydroquinase activity (1726) (Fig. 54). Conversely, aro-9 is used to select qa-1 and qa-2 mutants (1724, 1726). Homozygous crosses produce white ascospores.
aro-3 : aromatic-3
IIR. Between arg-5 (1%; 3%) and T(NM177)L, nuc-2 (347, 800, 1333). Not part of the aro-1 cluster gene.
Cloned and sequenced (872): Swissprot AROC_NEUCR, EMBL/Genbank U25818, PIR S11236.
Structural gene for chorismate synthase (EC 4.6.1.4) (704, 731) (Fig. 13). aro-3 is an unusual example of a chorismate synthase gene because it specifies a bifunctional polypeptide with both chorismate synthetase and flavin reductase activities (872). Requires a mixture of PABA, tyrosine, tryptophan, and phenylalanine for growth. Shows interallelic complementation (731). Leaky, giving hazy growth on minimal at 4 days, 34ºC; tests should be scored promptly (1546).
aro-4 : aromatic-4
Part of the aro-1 cluster gene. See aro-4 domain under aro-1.
aro-5 : aromatic-5
Part of the aro-1 cluster gene. See aro-5 domain under aro-1.
aro-6 : aromatic-6
VIL. Between ad-8 (8%; 12%), cpl-1 (5%) and lys-5 (3%) (822, 1815).
Structural gene for 3-deoxy-D-arabino-heptulosonic acid-7-phosphate synthase [DAHP synthase (Tyr), EC 4.1.2.15], one of the three isozymes inhibitable by tyrosine, phenylalanine, and tryptophan, respectively (Fig. 13). Grows on minimal medium except when both tryptophan and phenylalanine are present to inhibit the alternate synthases (822). Both activity-negative and allosteric-inhibition-negative alleles have been found (821). An aro-6 ad-8; aro-7 aro-8 stock is available (FGSC 4492).
aro-7 : aromatic-7
I. Between arg-1 (4%) and his-3 (l% or 2%) (822)
Structural gene for DAHP synthase (Phe) (EC 4.1.2.15), one of the three isozymes inhibitable by tyrosine, phenylalanine, and tryptophan, respectively (Fig. 13). Grows on minimal medium except when both tyrosine and tryptophan are present (822). Both activity-negative and allosteric-inhibition-negative alleles have been found (821). An aro-7 ad-3 aro-8; aro-6 stock is available (FGSC 4492).
aro-8 : aromatic-8
IR. Between so (7%; 11%) and R (4%) (822, 2129).
Purified and partially sequenced: Swissprot AROF_NEUCR; PIR P80576 (2178).
Structural gene for DAHP synthase (Trp) (EC 4.2.1.15), one of three isozymes inhibitable by tyrosine, phenylalanine, and tryptophan, respectively (Fig. 13). Grows on minimal medium except when both phenylalanine and tyrosine are present (822). Both activity-negative and allosteric-inhibition-negative alleles have been found (821). aro-7 aro-8 nic-1; aro-6 stocks are available (FGSC 4489, 4490).
aro-9 : aromatic-9
Part of the aro-1 cluster gene. See aro-9 domain under aro-1.
aro(p) : aromatic (polar)
Symbol used for polar mutants in the aro-1 cluster gene. See aro-l.
arom : aromatic
Symbol changed to aro.
arp1 : actin-related protein 1
Allelic with ro-4.
arp3 : actin-related protein 3
IIR. Linked near arg-12 (2092).
Cloned and sequenced: Swissprot ARP3_NEUCR, EMBL/GenBank U79737, Orbach-Sachs clone X7F05.
Encodes actin-related protein 3. Transcript levels decrease upon the induction of conidiation and then increase slightly during conidial differentiation (2092).
ars-1 : aryl sulfatase-1
VIIL. Between thi-3 (2%; 5%) and qa, Cen-VII, met-7 (l%), ile-1 (1%) (340, 1328, 1418).
Cloned and sequenced: EMBL/GenBank U89492; Orbach-Sachs clone X23G09.
Structural gene for aryl sulfatase (EC 3.1.6.1) (1329); see Fig. 24. Scored by color reaction with p-nitrophenyl sulfate. Because the enzyme is repressed in the wild type by traces of inorganic sulfate and other compounds present in all normal agars, screening on plates is carried out in an eth-lr cys-11 background, in which ars+ colonies have detectable, derepressed activity (1328). Scoring in crosses does not require this special background if the germinated spores are grown with cysteic acid (lmM) as the sole sulfur source (MgCl2 replacing MgSO4). Scoring method (1418). Reversion (1330). Mutants lacking aryl sulfatase were first isolated by screening in Neurospora crassa (1328) and were later shown to be allelic with a "natural" aryl sulfataseless gene, introgressed into N. crassa from one isolate of Neurospora tetrasperma, and with a gene which produces an electrophoretic variant enzyme in another natural isolate of N. tetrasperma (1329). Positively activated by cys-3 (1517); regulation reviewed (1283) (Fig. 24). Integration of ars-1 DNA at transgenic sites can eliminate the need for positive activation of expression (2148).
asc : ascus development
This symbol has been used for recessive mutants that affect ascus (or ascospore) development (518, 519). Some are barren; others result in much ascospore abortion. Only those asc mutations that have been mapped are listed here. Five remaining unmapped recessive mutants complement each other and mei-1. The asc mutants described by A. M. DeLange were shown not to be mutagen sensitive (775). See also asd, and mei.
asc(DL95) : ascus development (DL95)
IVR.
Perhaps a mei-1 allele.
asc(DL243) : ascus development (DL243)
IVR.
Perhaps a mei-l allele.
asc(DL879) : ascus development (DL879)
II. Linked to arg-5 (3%) (518).
Recessive. Meiosis is impaired in homozygous crosses. Seventy percent of ascospores abort and the total ascospore number is reduced, as are pachytene pairing and recombination (519). Viable ascospores are usually disomic for one or more linkage groups. Not tested for allelism with mus-7, which is homozygous-barren and maps in the same region.
Asc(KH2A83) : Ascus development (KH2A83)
VR. Left of al-3; probably right of his-1 (701, 844).
Ascospore abortion is 10% to 95% in heterozygous crosses and 90% to 95% in homozygous crosses. The frequency of aneuploids is increased. Sensitive to UV, but not to MMS or histidine (701).
asco : ascospore maturation
Used for a lys-5 allele with an autonomously expressed ascospore-ripening defect (1986).
Ascospore Color Mutants
Mutant genes with autonomous effects on ascospore pigmentation include asco, bs, per, ws, cys-3, lys-5, and pan-2. For applications, see refs. (597), (999), (1138), (1431), (1618), and (1985). cys-3 may be best for demonstrating segregation patterns in asci (1546, 1676) (Fig. 25). Except for bs and per, ascospores that lack wild-type pigment fail to germinate. Brown bs ascospores are fully viable. The unpigmented ascospores of per germinate spontaneously without heat shock and are killed by exposure to 60ºC.
asd-1 : ascus development-1
VIL. Between nuo19.3 (5T/18 asci) and cmt (2 or 3T/18 asci), Bml (5 or 6T/18 asci) (1447).
Cloned and sequenced: EMBL/GenBank U70861. On a common cosmid (pSV50 12.7B) with asd-3.
ASD-1 is homologous with Aspergillus rhamnogalacturonase B, which cleaves pectin (1449). Identified as a gene expressed preferentially under conditions that induce the sexual cycle. Essential for sexual development, but vegetative growth is normal. Expression is very low in mat Am44. In crosses homozygous for RIP-induced asd-1 mutants (1449, 1450), perithecial beaks are short, asci are normal in size and number, the two meiotic divisions are normal, but following the third division the distribution of nuclei is highly abnormal, ascospore delimitation fails, and no ascospores are produced (1450, 1673). Called sdv-10.
asd-2 : ascus development-2
VIIL. Linked to nic-3 (15%), ars-1 (36%) (1450).
Sexual development is blocked in homozygous mutant crosses. Perithecia are small, without beaks. No asci are formed, or only a few very short asci (1450, 1673). The mutation was probably caused by ectopic integration of nonhomologous transforming DNA into asd-2+ (1450).
asd-3 : ascus development-3
VIL. Between nuo19.3 (5T/18 asci) and cmt (2 or 3T/18 asci), Bml (5 or 6T/18 asci) (1447).
Cloned and sequenced: EMBL/GenBank AF136235. On the same cosmid (pSV50 12.7B) as asd-1.
ASD-3 is homologous to sugar-transporter proteins (1319). Identified as a gene expressed preferentially under conditions that induce the sexual cycle. RIP-mediated disruption of asd-3+ yielded strains resembling asd-1 mutants, with aberrant sexual development but normal vegetative growth. In homozygous mutant crosses, early sexual development is normal and many asci are formed, but ascospores are never delineated. Originally called sdv-15.
Asd-4 : Ascus development-4
IVR. Near met-5 (1277).
Cloned and sequenced (626a, 1277).
Encodes a GATA-type zinc finger DNA-binding protein unlike previously known GATA factors NIT-2, WC-1, WC-2, and SRE. Prevents development of asci. No detectable vegetative phenotype. Dominant. Obtained by RIP. The effect is reversed by introduction of the wild-type gene Asd-4+. Called BF99 (626a, 1277).
Asm-1 : Ascospore maturation-1
VR. Linked to al-3 (0T/18 asci) (49, 1447).
Cloned and sequenced: EMBL/GenBank U51117; Orbach-Sachs clones X17A07, X19C05.
A key regulatory gene involved in sexual development. Contains a conserved DNA-binding domain (49). The abundant product is nucleus-localized. The deletion mutant has a complex phenotype with vegetative defects recessive and sexual-phase defects ascus-dominant. Matted mycelium, stunted aerial hyphae, slow conidial germination, no protoperithecia. In crosses heterozygous for a deletion of Asm-1 (Asm-1+ x DAsm), all eight ascospores fail to mature in a large majority of asci. This is due to failure of transvection. Ascospore maturation depends on close meiotic pairing of Asm+ with its homologous allele (48). For other genes with ascus-dominant effects, see Ban, eat-2, Fsp, Pad, pkD, Prf and R.
asn : asparagine
VR. Between inv (4%; 9%) and pyr-6 (6%), pl (1%; 9%) (321, 322, 1383, 2014).
Requires asparagine for growth; does not respond to aspartic acid (2048). Lacks asparagine synthetase, (EC 6.3.5.4). Complementation between alleles (1240). asn auxotrophs appear dominant in heterokaryons when forced with ad, but recessive when forced with nic. May be inhibited by histidine (1546). Temperature-sensitive alleles T51M147 and T51M158 were originally classed as un mutants because growth is inhibited on organic complete medium. Symbol changed from asp (1579).
asp : aspartate
VR. Between at (0%; 3%) and per-1 (16%; 26%) (1582, 1585, 1604).
Growth is aided by aspartate or glutamate. Also some response to homoserine or leucine (556). Grows adaptively on minimal medium; adapting more rapidly at 25ºC than at 36ºC. Inhibited by alanine; 0.5 mg/ml alanine in test medium aids scoring. Symbol changed from aspt. The symbol asp was formerly used for asparagine. In 1973, new symbols were adopted to conform to bacterial usage for amino acid auxotrophs (1579), with asn used for asparagine and asp for aspartate.
aspt : aspartate
Symbol changed to asp.
at : attenuated
V. Linked to Cen-V (0T/57 asci) (1602). Between lys-1 (2%; 10%), cyt-9 (2%, 5%) and asp (0%; 3%) (1582, 1604).
Conidia are formed in small flecks or granular clumps on an agar surface, especially in a crescent at the top of a slant (1604) (Fig. 14). Growth and pigmentation are slower than wild type, but the surface of a slant is covered. A good marker most easily scored on minimal medium, where conidiation is less profuse than on complete. Allele NM221t is expressed better at 34ºC than at 25ºC. Called morph(M111) (1604).

FIGURE 14 The mutant at: attenuated, showing the formation of conidia in aggregates. Clumping of conidia can be seen macroscopically. Bar length 0.1 mm. Scanning EM photograph from M. L. Springer.
atp : ATPase
F-type ATPases consist of two major components, the catalytic core (F1) and the membrane proton channel (F0). F1 is composed of five polypeptide subunits, a, b, g, d and e, which are all nuclear-encoded. F0 has three subunits – a, b, and c; a and b are encoded in the mitochondrion and c in the nucleus. For a review, see ref. (208).
atp-1 : ATPase-1
I or V. Determined by blotting chromosomal DNA on CHEF gels.
Cloned and sequenced: Swissprot ATPA_NEUCR, EMBL/Genbank M84191, PIR JC1111, EMBL NCATPA, GenBank NEUATPA; EST SC1H4.
Structural gene for the ATP synthase a-subunit of the catalytic core F1, mitochondrial precursor (EC 3.6.1.34). This subunit contains a nucleotide-binding site, but the bound nucleotide is not hydrolyzed during the catalytic cycle (212).
atp-2 : ATPase-2
II. Linked to Cen-II, arg-5 (0T/18 asci) (1447).
Cloned and sequenced: Swissprot ATPB_NEUCR, EMBL/GenBank M84192, PIR JC1112, EMBL NCATPB, GenBank NEUATPB, NCF1ATPB; EST NP6B1.
Structural gene for ATP synthase b-subunit of the catalytic core F1, mitochondrial precursor (EC 3.6.1.34). The site of ATP synthesis is largely formed by this polypeptide (with a smaller contribution from the a-subunit) (212).
atp-9 : ATPase-9
Used for oli.
atr-1 : aminotriazole resistant-1
IL. Between suc (0/39), In(H4250)L and T(39311)R (1582, 1603).
Resistant to 0.5 mg/ml 3-AT in solid medium (added before autoclaving). Not resistant to acriflavine. Abnormal vegetative morphology. Female sterile. Resistance is recessive in heterozygous duplications from T(39311) (1582). Histidine in test media neutralizes the toxicity of 3-AT to wild type [3-AT affects histidine biosynthesis (1768)]. Resistance to 3-AT is also shown by acr-2 (931), leu-1, and leu-2 (1048, 1582).
aur : aurescent
The name originally used for al-1 allele 34508, which becomes visibly pigmented only in mature peripheral conidia and conidiophores. See al-1.
aza : azapurine resistant
Wild type is resistant to azapurines. A cpc-1 strain, which is sensitive, was used for selecting azapurine- resistant mutants. Three resistance loci are known. aza-1 and aza-2 mutants have been lost, but their described characteristics and map locations should enable recurrences to be recognized. They were selected using aza-adenine, but at least one allele at each locus also showed resistance to azaguanine. aza-3 was selected for resistance to azaguanine using a different procedure, and resistance to aza-adenine has not been determined.
aza-1 : azapurine resistant-1
IL. Left of mat (23%) (995).
Resistant to purine analogues 8-aza-adenine and 2-6-diaminopurine. (One allele of four was also resistant to 8-azaguanine.) Obtained in cpc-1 (mts), which is inhibited by the analogs. Selected and scored using l mg/ml 8-aza-adenine in medium. Resistance is recessive in a heterokaryon. At least one allele secretes purines. Called aza-adenine-resistant (995). Strains lost (991).
aza-2 : azapurine resistant-2
IL. Linked to mat (2%), aza-1 (39%) (995).
Resistant to purine analogs 8-aza-adenine and 8-azaguanine. Sensitive to 2-6-diaminopurine. Obtained and scored as described for aza-1. Resistance is recessive in a heterokaryon. Called aza-adenine-resistant (995). Strains lost (991).
aza-3 : azapurine resistant-3
III. Linked to trp-1 (14%) (884).
Resistant to purine analogs 8-azaguanine and 6-mercaptopurine. Obtained upon limiting adenine in an adenine auxotroph, which is inhibited by the analogs. Selected and scored using ad-3A ad-3B; ad-2 on medium with 2 mg adenine sulfate and 200 mg/ml azaguanine. Relative resistance of aza-3R and aza-3S to 8-aza-adenine not determined. Resistance is recessive in a heterokaryon. Hypoxanthine can be used as the sole purine supply (883, 884). Called azaguanine-resistant.
azs : azide sensitive
Unmapped. Unlinked to has (595).
Cannot produce the inducible azide-sensitive respiratory pathway when grown in the presence of chloramphenicol (595). Obtained from the double-mutant strain ANT-l (antimycin-sensitive), which was also the source of has (588, 591, 595). The double mutant azs; has has been used to obtain oligomycin-resistant mutants (596) and mutants deficient for succinic dehydrogenase (590).
Return to the 2000 Neurospora compendium main page