
Bm : mauve
IL. Left of nit-2 (30%) (739).
Colonies are mauve on special dye medium where the wild type is blue (739).
bal : balloon
II. Between T(AR179) (hence of thr-2, thr-3) and T(ALS176), arg-5 (1%; 7%) (1578, 1580, 1582, 1585). Probably left of Cen-II (714, 800).
Forms a smooth, slow-growing hemispherical colony (1548). Fully female-fertile, which is uncommon for colonial mutants having such restricted growth. Recessive in heterokaryons. Glucose-6-phosphate dehydrogenase deficiency (1853, 1854, 1857). (col-2 and fr are also deficient in G6PD.) Reduced levels of NADPH (233), linolenate (241), and peptides in the cell wall (2249). Morphology is subject to alteration by modifiers that are commonly present in laboratory stocks, resulting in spreading growth and conidiation. See su(bal). Photograph (235, 1851, 1853). Allele C-1405 called mel-2 (1409, 1585).
Ban : Banana
IL. Left of mat (14%), Probably left of leu-3 (1684).
Each ascus delimits a single giant ascospore enclosing all four meiotic products and their mitotic derivatives (Fig. 15). The ascus trait is dominant and almost completely penetrant. The mature giant ascospores are capable of germination and usually give rise to mixed cultures. The giant ascospore in Ban is formed not by hardening of the ascus wall, as in Iasc, but by hardening of the ascus membrane within a normal wall, which it may not fill completely. Vegetative defects are recessive. Morphology is abnormal, with short aerial hyphae and no protoperithecia (1684). In older perithecia, the prefusion nuclei in the croziers revert to mitosis, which is synchronized and favorable for cytological observation (Fig. 16). (Croziers in Prf show similar behavior.) Ban has been used to rescue segregants that would not survive as homokaryotic haploids (1674). Ban arose in a burst of spontaneous mutants in a cross involving mei-3, which is also located in IL (1469). Some Ban stocks may contain mei-3 or a mei-3 partial revertant that was recovered from the same cross.

FIGURE 15 The mutant Ban: Banana. A maturing giant ascospore from Ban+ x Ban, showing striations throughout its length. Sixteen nuclei can be seen before the ascus wall becomes opaque. Photograph by N. B. Raju. From ref. (1684 ).
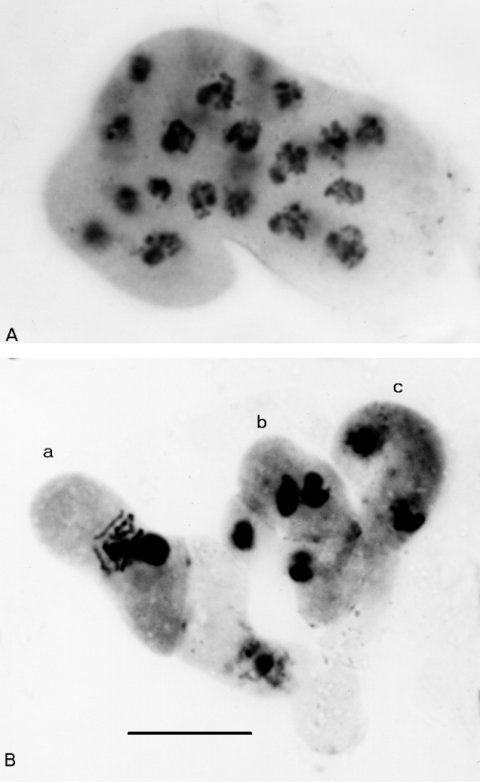
FIGURE 16 (A) multinucleate crozier cyst from Ban+ x Ban. Rather than fusing, the haploid pronuclei have reverted to dividing by mitosis in rounds that are synchronous. (B) Normal crozier development in the wild type, for comparison. (a) A young ascus following fusion of the haploid parental nuclei. (b) A three-celled crozier with a binucleate penultimate cell, which will give rise to an ascus. (c) A crozier with the two nuclei in conjugate division. X1500. Stained with haemotoxylin. Photographs by N. B. Raju, from ref. (1684 ).
bas : basic amino acid transport
Probably allelic with pmb. Also called basa.
bat : basic amino acid transport
bd : band
IVR. Between met-5 (9%) and nit-3 (19%) (1185, 1794).
Because dense bands of conidia are produced on appropriate solid medium at intervals of about 24 hr (1792, 1794), the mutant has been used extensively to study circadian rhythms (239, 560, 621, 1382, 1794). bd has no effect on the underlying clock mechanism, but allows the visible expression of rhythm (622). Grow rate is about 70% that of the wild type (232). Conidiation is enhanced, even on slants (232). CO2 inhibits conidiation and, thus, inhibits banding; bd is much less sensitive than wild type to this effect of CO2 (1792). Biotin starvation leads to a phenocopy in wild type and to increased persistence of banding in bd (2206). Originally identified in a bd; inv strain called "timex" (1791). bd alone is sufficient to cause banding (1794). Used to study conidiation under nonstarvation conditions (1820). Used in a study of morphological differentiation patterns such as concentric rings and radial zonations (528). Expression is affected by changing the concentrations of agar, sugar, and salts. Conidial scatter is eliminated in the double mutant bd; csp (239). Conveniently scored by conidial banding on agar in long tubes or large plates at 25oC in constant dark or in a dark-light cycle, but not in constant light (1791).
ben : benomyl-resistant
Symbol changed to Bml.
Beta-tubulin
See Bml.
bis : biscuit
Changed to pk. See p. 270 of ref. (1592).
bld : bald
IVR. Right of pdx-1 (4%) (1592).
Aerial hyphae at the top of a slant are devoid of conidia. More extreme at 25°C than at 37°C, where powdery conidia are formed in a zone below the top. Called mo(NM213t) and incorrectly stated to be wild type at 34°C (1592).
ble : bleomycin resistant
Introduced from transposon Tn5.
This gene from Tn5, which confers resistance to bleomycin and phleomycin, has been placed under control of the Neurospora am promoter and used as a dominant selectable marker for Neurospora transformation (74). See also amdS.
bli-3 : blue light induced-3
IVR. Right of trp-4 (1T/15 asci). Linked to Fsr-51 (0T/15 asci), Fsr-13, vma-8 (0T/18 asci) (1447).
Cloned and sequenced: Swissprot BLI3_NEUCR, EMBL/GenBank X81318, GenBank NCBLI3.
Increased bli-3 mRNA level is detected after 2-min exposure of a dark-grown culture to blue light. Obtained as a cDNA clone by differential screening, using probes from illuminated mycelia and dark controls (1946).
bli-4 : blue light induced-4
IIR. Linked to Fsr-34 (0T/18 asci), between eas (1T/18 asci) and Fsr-17 (5T, 1NDP/18 asci), leu-6 (9T, 1NPD/ 18 asci) (1447).
Cloned and sequenced: Swissprot BLI4_NEUCR, EMBL/GenBank X89499, GenBank NCBLI4GEN.
Encodes a blue-light-induced, short-chain alcohol dehydrogenase (1530). An increased bli-4 mRNA level is detected after 2-min exposure of a dark-grown culture to blue light.
bli-7 : blue light induced-7
Allelic with eas.
bli-13 : blue light induced-13
Unmapped.
Also strongly induced by continuous white light (1946).
Bml : Benomyl resistant
VIL. Between cys-2 (2%), cpc-1 and ad-1, Cen-VI. Linked to ylo-1 (2% or 3%) and un-13 (1/64), perhaps to the right (197, 1582, 1815).
Cloned and sequenced: Swissprot TBB_NEUCR, EMBL/GenBank M13630, PIR A25377, GenBank NEUTUBB; EST NC3D10; pSV50 clone 24:11A, Orbach-Sachs clones X15H04, X18D12, G12C10, G14C11, G15C03, G15E01.
Encodes the b-subunit of tubulin. Mutation causes semidominant resistance to benomyl (197), a benzimidazole fungicide that affects hyphal tip extension and polarity. Mutations showing different degrees of resistance to benomyl all map to the Bml locus (2095). Typically, 0.5-1 ug/ml benomyl inhibits the wild type but not the mutant. Benomyl causes multiple germ-tube emergence from germinating macroconidia (268) and affects the level of Bml mRNA during germination (882). A Phe-to- Tyr change at position 167 confers resistance in the original mutant, 511 (1503). A Glu-to-Gly change at position 198 confers both resistance to the related benzimidazole fungicide carbendazim and sensitivity to diethofencarb, an N-phenylcarbamate fungicide to which the wild type is normally resistant (696). Additional amino acid changes can result in simultaneous resistance to both fungicides (694, 695). Bml has been used as a dominant selectable marker to introduce unselected genes by transformation (1503, 2167). Its use has the disadvantage, however, that introduced genes cannot be recovered efficiently in the progeny when these transformants are crossed, presumably because duplicate b-tubulin-encoding sequences in the transformants are subject to inactivation by RIP, causing the progeny to be inviable. Consequently, hph has seen wider use as a dominant selectable marker for transformation (1502, 1504, 1977). Bml has been called Ben (197); mbic (931), and tub-2 (1503).
bn : button
VII. Linked to met-7 (0/93), wc-1 (1%) (1585). Right of T(T54M50), hence, of thi-3 (2%) (1548, 1578).
Nonconidiating, restricted colonial growth (102, 1548). Altered 6-phosphogluconate dehydrogenase (EC 1.1.1.44) (1852, 1854). (6PGD is also affected by col-10.) Levels of NADPH (233) and linolenic acid (241) are reduced. Used to study apical growth and polarization (2116). Germination may be better on minimal than on complex complete medium (1546). Allelic with col-3 and complements col-2 in heterokaryons (1565).
bs-1 : brown ascospore-l
IR. Linked to un-1 (9%), probably to the right (1603).
Ascospores are brown rather than black at maturity. Viability is not impaired. Expressed autonomously, allowing visual scoring in heterozygous asci (1603). Used to study factors affecting second-division segregation frequencies (1138). Translocation T(NM139) bs has a similar phenotype but is not allelic with bs-l, although one breakpoint is in IR proximal to al-2 (1580).
Return to the 2000 Neurospora compendium main page