c:
Used in early publications as a symbol for het-c, cot-4, or cy
ca-1 : calcium transport-1
I. Linked to cys-9 (10%), un-1 (10%) (693).
Optimal growth is obtained in liquid medium containing low calcium (693). Alleles 19 and 31 grow optimally with 10 mM calcium in the medium (408, 693) and are defective in vacuolar calcium transport (408). Allele 101, which grows optimally at 100 mM calcium, results in a shortened circadian rhythm (693). The effects of calcium on growth are not observed on solid medium.
ca-3 : calcium transport-3
I. Linked to ca-1 (5%), cys-9, un-1 (10%) (693).
Optimal growth is obtained in liquid medium containing 10 mM calcium. The effect of calcium on growth is not observed on solid medium (693).
ca(23-2) : calcium transport (23-2)
Unmapped.
A long lag phase is seen in medium containing low calcium. Phase of the circadian rhythm is shifted by temperature but not by light (1491).
caf-1 : caffeine resistant
VL. Between dgr-1 (8%), T(AR30) (19%) and mus-16 (4%), T(AR33) (5%; 12%), lys-1 (4%; 14%) (926, 1578, 1582, 1598, 1602).
Resistant to caffeine (927). Resistance is dominant in duplications from T(AR33) (1598). Scoring is clear at 25ºC and poor at 34ºC, on slants with 2.5 mg/ml caffeine in minimal medium without sorbose (1322). Also readily scorable using conidial suspensions spotted on plates containing 2 mg/ml caffeine in minimal sorbose medium (1582).
can : canavanine resistant
Changed to cnr.
car : carbohydrate
IVL. Linked to cys-10 (1%) (815).
Affects carbohydrate transport. Altered morphological rhythm is associated with a deficiency in the low-affinity glucose transport system. On glucose, car produces dense and sparse mycelium in cycles (period about 50 hr). On acetate, car is insensitive to the light-dark cycle and has a normal conidiation cycle with period about 24 hr. The cycle is not circadian. Periodicity is affected by the composition of the medium rather than by time. The car mutant originated from a cross between pat and acu-7, but acu-7 is not necessary for expression of the phenotype. Called Lpcar: long period carbohydrate (815). The symbol car has also been used for some carotenoid mutants, at least one of which is an al-2 allele (2019), and for a carbohydrate mutant (2007) that is not available for testing.
cat : catalase
Expression of the three structural genes is regulated differentially during development and in response to heat shock and treatment with oxidants. Products of the three genes differ in electrophoretic mobility and molecular weight (366, 1917).
cat-1 : catalase-1
IIIR. Linked to T(1), ylo-1 (1/26), trp-1 (17%) (1447, 1917).
Cloned and partially sequenced: EMBL/GenBank AI392552; EST SC3A3.
Encodes major catalase enzyme (EC 1.11.1.6) predominant in growing mycelium (366). Used to study the effects of singlet oxygen on CAT-1 heme (1216).
cat-2 : catalase-2
VIIR. Probably linked to for, frq (£10T + 0NPD/18 asci), cox-8 (£6T + 1NPD/18 asci) (1447, 1917).
Cloned and partially sequenced: GenBank AA901787; EST NM 10 E2.
Structural gene for minor catalase enzyme (EC 1.11.1.6) predominant in mycelium after heat shock (366, 1917).
cat-3 : catalase-3
IIIR. Between Fsr-45 (2T, 1NPD/18 asci) and Tel-IIIR (5T/18 asci) (1447).
Cloned and partially sequenced: EST EMBL/GenBank AA901970; EST NM6H12.
Structural gene for minor catalase enzyme (EC 1.11.1.6) predominant in mature conidia (366).
cax : calcium exchanger
VIL. Linked to Tel-IVL (3T/18 asci) (1447).
Cloned and sequenced: EMBL/GenBank AF053229; Orbach-Sachs clones X5D07, G3A09, G23H08.
Encodes a calcium transport protein in the vacuolar membrane. The predicted polypeptide contains 11 membrane-spanning helices. cax mutants grow normally (1272).
cbh-1 : cellobiohydrolase-1
Unmapped.
Cloned and sequenced: Swissprot GUX1_NEUCR, EMBL/GenBank X77778, GenBank NCCBH1, PIR S42093.
Structural gene for the major 1,4-b-cellobiohydrolase (exoglucanase, exocellobiohydrolase) (EC 3.2.1.91), a component of the cellulase complex.
ccb-1 : constitutive carotenoid biosynthesis-1
Unmapped.
Mycelia in dark-grown liquid cultures become deep orange, but the mRNA levels of light-regulated genes do not increase (1206, 1208). Carotenoid synthesis after 2 days of growth in the dark resembles that of light-induced wild type. mRNA levels of carotenoid biosynthetic genes are increased following exposure to blue light, but those of developmentally regulated genes expressed during conidiation (con-8, con-10, and eas) are not. Conidia are formed in liquid medium (1208). Recessive. Obtained by selection using an al-3-promoter::mtr fusion strain in which mtr-mediated uptake of tryptophan and p-fluorophenylalanine depends on light induction (286).
ccb-2 : constitutive carotenoid biosynthesis-2
Unmapped.
Mycelia in dark-grown liquid cultures become deep orange, but the mRNA levels of light-regulated genes do not increase (1206, 1208). Poor growth and conidiation. Hyphae are short, with chains of separated cells. mRNA level for the developmentally regulated gene eas is increased in response to blue light, but mRNAs for carotenoid genes show little or no increase. Attempts to cross or to form heterokaryons have been unsuccessful (1208). Origin same as for ccb-1.
ccg-1 : clock-controlled gene-1
VR. Between Cen-V (4T/18 asci) and ilv-2 (4T/13 asci) (1447).
Cloned and sequenced: Swissprot GRG1_NEUCR; EMBL/GenBank X14801, EMBL NCMORCCG, GenBank NEUMORCCG; pSV50 clone 29:6C.
Encodes a highly abundant transcript that peaks early in the circadian morning (147, 1226). Regulated by development and light (61, 1206); light-regulation normally requires wc-1 and wc-2, but the cot-1 mutation suppresses the requirement for wc-2 (61). A ccg-1::tyrosinase reporter gene has been used to screen for mutants with altered expression of conidiation-related genes (1093). Allelic with grg-1, which was isolated on the basis of regulation by glucose (575, 1315, 2055, 2186).
ccg-2 : clock-controlled gene-2
Allelic with eas.
ccg-4 : clock-controlled gene-4
IL. Between mat (3T/18 asci; 0/18) and Cen-I (2T/18 asci) (1447).
Cloned and sequenced: EMBL/GenBank AF088909, U46085, X81318; EST NM9C1.
The transcript level peaks early in the circadian morning and is regulated by both light and developmental signals (147). The ccg-4 sequence shows similarity to a mating-type-specific pheromone precursor gene from Cryphonectria parasitica and Magnaporthe grisea, including multiple repeats of a putative pheromone sequence bordered by KEX2 processing sites. Consistent with mating-type-specific pheromone activity, the expression of ccg-4 is restricted to cells of mating type A (189). See eat-1, eat-2, mfa-1, and scp for other genes that are expressed differentially in the two mating types.
ccg-6 : clock-controlled gene-6
VR. Linked to al-3, inl (0 or 1T/18 asci) (1447).
Cloned and sequenced: Swissprot CCG6_NEUCR, EMBL/GenBank U46086, AF088908; EST NC5G12.
RNA level peaks late in the circadian night and is regulated by both light and developmental signals (147).
ccg-7 : clock-controlled gene-7
Allelic with gpd-1.
ccg-8 : clock-controlled gene-8
V. Linked to lys-1, Cen-V (0T/18 asci) (147, 1447).
Cloned and sequenced: Swissprot CCG8_NEUCR, EMBL/GenBank U46087, AF088907.
The mRNA level peaks late in the circadian night but is not regulated by either light or developmental signals (144, 147).
ccg-9 : clock-controlled gene-9
VIIL. Linked to nic-3 (2T/18 asci) (1447).
Cloned and sequenced: EMBL/GenBank U46088, AF088906.
Transcript peaks late in the circadian night and is regulated by light and developmental signals (147). Sequence comparisons suggest that ccg-9 encodes trehalose synthase (1909).
ccg-12 : clock-controlled gene-12
Allelic with cmt.
cdr-1 : cadmium resistant-1
II. Left of arg-12 (35%) (1182).
Resistant to 10-20 mM cadmium chloride on solid medium, but similar to wild type when grown immersed in liquid medium. Not cross-resistant to zinc, copper, or cobalt. Not osmotic-sensitive. Distinctive morphology (1182). Stock lost (1277).
cdr-2 : cadmium resistant-2
VIIL. Between nic-3 and met-7 (1182).
Similar to cdr-1 in resistance, but morphologically distinct (1182). Stock lost (1277).
cdr-3 : cadmium resistant-3
Unmapped. Not in II or VII (1182).
Present in Neurospora intermedia strain P3 (FGSC 1768), which originated from a volcanic area of hot springs and vents in Unzen, Japan. Similar to cdr-1, but morphologically distinct (1182).
cel-1 : chain elongation-1
IVR. Linked to pan-1 (1%), cot-1 (0/17) (1585).
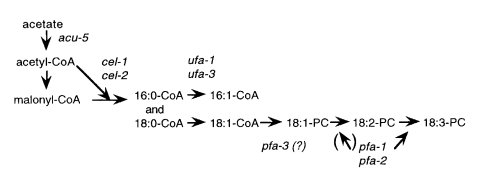
Impaired synthesis of saturated fatty acids attributed to defective fatty acid chain elongation (871). Defective fatty acid synthase complex (603) (Fig. 17). Activity of the α-subunit of fatty acid synthase is reduced to 1.2% of wild type. Growth is normal with palmitate as supplement (239). Tween 40 provides a convenient supplement. The requirement is leaky at 22ºC (603), but not at 34ºC (1585). Sensitivity to oligomycin is increased [L. R. Forman and S. Brody, cited in ref. (536)]. Used to change fatty acid composition (237) and make the circadian clock sensitive to fatty acids and temperature (239, 1294). Effects on the circadian clock are reviewed in ref. (1130). Temperature compensation of the clock is lacking in cel-1 (1295). Used to study membrane lipid-phase transitions and electrical properties (679, 680). Used to incorporate photolabile azido fatty acid probes for membrane studies (342). Palmitate is used more efficiently in cel-1 than in wild type for the synthesis of unsaturated fatty acids (754). Called ol: oleic (1585) and fas: fatty acid synthesis (603). The initial report of oleic utilization (1585) was incorrect (871), probably because of impurities.

FIGURE 17 The biosynthetic pathway for the major fatty acids of Neurospora, showing the sites of action for the cel, ufa, and pfa genes. Abbreviations for the fatty acids are as follows: 16:0, palmitate; 16:1, palmitoleate; 18:0, stearate; 18:1, oleate; 18:2, linoleate; 18:3, linolenate. The fatty acids generally are found esterified to complex lipids such as phosphatidylcholine (PC). The genes ufa-2 and ufa-4 have pleiotropic effects and may affect auxiliary electron transport reactions. The genes pfa-4 and pfa-5 appear to affect the pathway of biosynthesis of 18:3 indirectly, as do bal, fr, and col-2. Original figure from Marta Goodrich-Tanrikulu.
cel-2 : chain elongation-2
IVR. Linked to cot-1 (1%) (755), bd (232).
Impaired synthesis of saturated fatty acids (755) (Fig. 17). The β-subunit of fatty acid synthase is defective. Activity is lower than in cel-1, making cel-2 potentially more useful for enriching membranes in supplemental fatty acids. Revertible. Fertility is reduced. Growth is normal on palmitate (232). Palmitate is used more efficiently in cel-2 than in wild type for the synthesis of unsaturated fatty acids (1997).
cell-1 : cellobiase/cellulase
Unmapped. Segregates as a single gene, independent of gluc-1.
Both cellulase and cellobiase are produced constitutively. Levels of aryl-b-glucosidase are not affected. Recessive to wild type in heterokaryons (1420). Isolated using gluc-1 and selecting for high activity in destroying the b-glucoside esculin (582). (Cellulase, cellobiase, and aryl-b-glucosidase normally are induced simultaneously by cellobiose.)
cem-1 : condensing enzyme with mitochondrial function
Unmapped.
Cloned and sequenced: EMBL/GenBank AF021234.
A nuclear gene that encodes a mitochondrial protein, 3-oxyacyl (acyl carrier protein) synthase (EC 2.3.1.41) (250).
Cen : Centromere
Knowledge of the physical structure and organization of the centromeres is based on the only sequenced centromere region, Cen-VII (275, 340). The Neurospora centromere is structurally more similar to that of Drosophila than to that of Saccharomyces. If centromeres of the other Neurospora chromosomes are comparable in size and content, centromeric repeats may make up as much as 7% of the Neurospora genome. A defective copia-like transposable element Tcen is associated exclusively with all seven centromere regions, which also contain other relic transposons and many defective Tad elements (275).
Several methods are available for the genetic mapping of centromeres relative to flanking gene loci: (i) tetrad analysis with physically ordered asci (1199); (ii) analysis of unordered asci in crosses heterozygous for Spore killer (1602) or other known centromere markers; (iii) duplication coverage of a flanking gene in a segment carrying gene loci known to be located in a given arm (1566); and (iv) cotranslocation to another chromosome arm of a flanking gene together with other loci whose arm is known. The third method is least laborious when appropriate rearrangements are available. Linkage group arms were first defined as left (L) and right (R) by ref. (103), using mat, pe, ser-l, pdx-l, ilv-l, rib-l, and nt as reference markers.
Cen-I : Centromere-I
I. Between arg-3 (2%), T(39311)R and sn (1/271 second-division segregation), os-4, T(AR173L), his-2 (733, 1578, 1580, 1605, 1963).
The amount of crossing over per unit of physical distance is much lower near Cen-I than in more distal regions of I or in the overall genome (1299, 1740). Recombination between arg-3 and his-2 varies from 3% to 18%, depending on rec genotype (338).
Cen-II : Centromere-II
II. Between T(AR179)R and T(ALS176), arg-5 (0T/18 asci) (1447, 1578, 1580) (bal also lies between these translocations). Ordered asci indicate that Cen-II is probably between bal and arg-5 (714, 800).
Cen-III : Centromere-III
III. Between acr-7 (2T/18 asci) and thi-4 (1T/18 asci), sc (4T/230 asci). Linked to crp-2 (0T/18 asci) (103, 1447). Order relative to acr-2 is uncertain.
Cen-IV : Centromere-IV
IV. Right of ace-4, cut, fi by inference from recombination distances. Left of psi [D. R. Stadler, A. M. Towe, and M. Loo, cited in ref. (1220)], T(ALS159) (1580).
Cen-V : Centromere-V
V. Between lys-1 (5%), cyt-9 (2%), T(MB67)R and asp. No recombination with at (0/57 asci) (1578, 1602).
Cen-VI : Centromere-VI
VI. Between ad-1 (l% or 2%) and T(AR209), rib-1 (l%; 4%), pan-2 (2%) (296, 1580, 1986). glp-4 is also between ad-1 and rib-1 (2160).
Cen-VII : Centromere-VII
VII. Between ars-1, qa cluster and met-7 (l%) (340).
Cloned and sequenced: EMBL/GenBank L23168, AF079510, GenBank NEURSCENTR.
The 450-kb region that contains Cen-VII is recombination-deficient and A + T-rich relative to flanking regions. The region contains repetitive sequences which hybridize to sequences that map at or near the centromeres of all the other linkage groups. The repeated sequences are divergent, with differences that are predominantly GC ® AT transitions, suggesting that they have been subjected to RIP. Recombination frequencies relative to physical length decline dramatically in this region, which comprises 11% of the linkage group VII DNA (340). Reduction extends into the arms, gradually diminishing in the 200-bp interval Cen-VII-wc-1 [G. Macino, cited in ref. (340)]. Sequencing of a 16-kb portion revealed a tightly nested cluster of simple sequence repeats and degenerate retrotransposon-like elements —Tcen, which is copia-like and is found exclusively in centromere regions, Tgl1, identified as gypsy-like by sequence resemblance to the Ty3-2 transposon of Saccharomyces, and Tgl2, another immobile putative gypsy-like element. Multiple degenerate fragments of the Line-like Tad element also are present. The defective transposons all contain transition mutations typical of RIP. Multiple clusters of similar content are present elsewhere in the Cen-VII region (275).
cfp : cellular filament polypeptide
VIIR. Between for (1 or 2T/18 asci) and T(5936) (810, 1447). In a cosmid adjoining that carrying ras-3 (1267).
Cloned and sequenced: Swissprot DCPY_NEUCR, EMBL/GenBank L09125, U56927, PIR JN0782, EMBL NCCFPX, GenBank NEUCFP; EST NC3D2; Orbach-Sachs cosmid G15:G6.
Structural gene for pyruvate decarboxylase (EC 4.1.1.1), which forms 8- to 10-nm cytoplasmic filaments (1739). These filaments are abnormally sized and shaped in strains mutant at the sn locus (34, 40, 1739). Identified independently and called pdc-1 (1267).
cfs(OY305) : caffeine sensitive(OY305)
IR. Between mat and al-2 (2279).
Growth is inhibited by caffeine (0.2 mg/ml) and stimulated by adenine. Morphologically abnormal. Not sensitive to caffeine in the presence of adenine. Growth is slow on minimal medium. Probably UV-sensitive by spot test. Not tested for allelism with ad-3A, ad-3B, or ad-9 (2279).
cfs(OY306) : caffeine sensitive(OY306)
IR. Near al-2, probably to the right (2279).
Growth is inhibited by caffeine (0.2 mg/ml) and by adenine. Morphologically abnormal. Grows slowly on minimal medium and is not stimulated on complete medium. Complements cfs(OY305), cfs(OY307) (2279).
cfs(OY307) : caffeine sensitive(OY307)
IR. Between mat and al-2. Near cfs(OY305) (2279).
Growth is inhibited by caffeine (0.2 mg/ml) and by adenine. Morphologically abnormal. Grows slowly on minimal medium and is not stimulated on complete medium. Complements cfs(OY305), cfs(OY306) (2279)
chol : choline
There are four identified auxotrophic genes that respond to choline. The catalytic activity of the first is phosphatidylethanolamine N-methyltransferase (EC 2.1.1.17). The product of the second gene, methylene-fatty-acyl-phospholipid synthase (EC 2.1.1.16), catalyzes the second and third methylations in the pathway of phosphatidylcholine biosynthesis from phosphatidylethanolamine (Fig. 18). Two of the chol genes are known to be involved in the synthesis of phosphatidylcholine.
chol-1 : choline-1
IVR. Linked to ad-6 (l%), probably to the right (1255).
Requires choline (905). Also uses mono- or dimethylaminoethanol (892) (Fig. 18). Deficient in S-adenosylmethionine phosphatidylethanolamine N-methyl transferase (EC 2.1.1.17) (430, 1806), the enzyme catalyzing the first step in the phosphatidylcholine (lecithin) biosynthetic pathway. Morphology on limiting choline is abnormal, colonial (430). Colonies from single conidia on minimal agar medium resemble inhibited mat-A/mat-a duplications, with swollen hyphae and dark pigment in the presence of phenylalanine + tyrosine (1546). Phospholipid composition is abnormal on limited concentrations of supplement (934). Best scored late on minimal medium, where the mutant grows slightly and then stops (1546). Used to study the inhibition of cytochrome-mediated respiration and of conidiation when lecithin is depleted by choline starvation (1006, 1007). Used to demonstrate that mitochondria reproduce by division (1233). The chol-1 mutation lengthens the period of circadian rhythm of conidiation. In the chol-1; frq double mutant, the period is lengthened to over 50 hr on low choline. In combination with a null frq mutation, chol-1 remains rhythmic on low choline (1131).
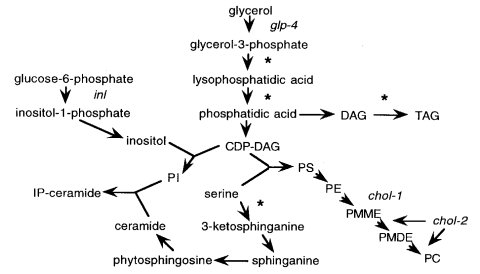
FIGURE 18 The biosynthetic pathway for the major fatty acid-derived lipids. Sites where fatty acids are incorporated are marked with asterisks. The major phospholipids are phosphatidylserine (PS), phosphatidylethanolamine (PE), phosphatidylcholine (PC), and phosphatidylinositol (PI). The sites of action of the inl and chol genes are shown. Abbreviations: DAG, diacylglycerol.; TAG, triacylglycerol; IP-ceramide, inositolphosphorylceramide. The PC intermediates phosphatidylmonomethylethanolamine (PMME) and phosphatidyldimethylethanolamine (PMDE) do not accumulate in wild type. Minor reactions occur, such as the phospholipase-mediated conversion of PC to DAG and the conversion of DAG to PC, which occurs in chol mutants supplemented with choline. For simplicity, these are not illustrated. Sites of action of the chol-3, chol-4, and un-17 genes are not known. Original figure from Marta Goodrich-Tanrikulu.
chol-2 : choline-2
VIL. Between Tip-VIL and nit-6 (6%; 8%) (1582, 1585).
Requires choline (895). Also uses di- but not monomethylaminoethanol (892) (Fig. 18). Deficient in methylene-fatty-acyl-phospholipid synthase (EC 2.1.1.16), catalyzing steps 2 and 3 in the phosphatidylcholine (lecithin) biosynthetic pathway. The only allele, 47904t, is leaky on minimal medium at 22ºC, but not at 34ºC (934). Phospholipid composition is abnormal on limiting choline (934). Growth is colonial on limiting supplement at 34ºC and on minimal at 25ºC. Resistant to aminopterin.
chol-3 : choline-3
VR. Between at (18%) and al-3 (19%; 47%) (1582, 1593).
Requires choline, but growth is subnormal even on optimal supplement (1593). The mutant was obtained and the requirement identified in the E. L. Tatum laboratory.
chol-4 : choline-4
IV. Linked to cot-1 (32%), chol-1 (3 prototrophs among 24 intercross progeny) (1582, 1593).
Responds to choline, but growth is subnormal even on optimal supplement (1593). The mutant was obtained and the requirement identified in the E. L. Tatum laboratory (1593).
chr : chrono
VI. Between chol-2 (10%) and pan-2 (712).
Altered period of circadian conidiation rhythm. The one known allele is incompletely dominant, with a 23.5-hr period at 25ºC in csp+ background. Temperature compensation is good above 30ºC (711, 713).
chs-1 : chitin synthase-1
V. Between rDNA (5/18), con-4a (1/18) and am (4/18), chs-3 (5/18) (1447).
Cloned and sequenced: Swissprot CHS3_NEUCR, EMBL/GenBank M73437, PIR E38192, EMBL NCCHT, GenBank NEUCHT.
Structural gene for chitin synthase 1 (EC 2.4.1.16) (chitin-UDP acetylglucosaminyl transferase 1). The RIP-induced mutant is slow-growing and morphologically abnormal (2269).
chs-2 : chitin synthase-2
IVR. Linked to nit-3 (6/18), Fsr-4 (0/17), uvs-2 (0/18) (1447).
Cloned and sequenced: Swissprot CHS2_NEUCR, EMBL/Genbank X77782, PIR B45189, EMBL NCCHITSYN, GenBank NCCHITSYN.
Structural gene for chitin synthase 2 (EC 2.4.1.16) (chitin-UDP acetylglucosaminyl transferase 2). RIP-induced mutant has no phenotype other than increased sensitivity to edifenphos (543).
chs-3 : chitin synthase-3
VR. Linked to am (1 or 2/18), inl (1 or 2/18) (1447).
Cloned and sequenced: Swissprot CHS3_NEUCR, EMBL M73437, GenBank AF127086, PIR A41638.
Structural gene for CHS-3 (EC 2.4.1.16) (chitin-UDP acetyl-glucosaminyl transferase 3), a class I chitin synthase, which is essential (175, 203).
chs-4 : chitin synthase-4
I. Linked to leu-4, mat, arg-3 (0/18) (1447).
Cloned and sequenced: Swissprot CHS4_NEUCR, EMBL/GenBank U25097, EMBL NC25097, PIR S61886.
Encodes the structural gene for chitin synthase 4 (EC 2.4.1.16) (chitin-UDP acetylglucosaminyl transferase 4) (542). RIP-induced mutant is abnormal only on sorbose (542).
chs-5 : chitin synthase-5
IVR. Between arg-14 (2/18) and cot-1 (1/18) (1447).
Cloned and partially sequenced (2267): GenBank A 1397637; EST SC5D3.
Structural gene for chitin synthase 5 (EC 2.4.1.16) (chitin-UDP acetylglucosaminyl transferase 5) (2267).
ci-1 : cycloheximide inducible-1
Unmapped.
Cloned and sequenced: EMBL/GenBank X15033, GenBank NCCI1, PIR S09643.
Encodes a 70-kDa polypeptide that is a member of the cytochrome P-450 family (69). Highly induced by the partial inhibition of protein synthesis resulting from low doses of cycloheximide or the depletion of polyamines; not induced by heat shock or starvation (24, 805).
cia35 : chaperone protein 35
Unmapped.
Cloned and sequenced: EMBL/GenBank AJ001726, GenBank NCCIA35.
Encodes a chaperone protein involved in the assembly of mitochondrial complex I (1105).
cia80 : chaperone protein 80
Unmapped.
Cloned and sequenced: EMBL/GenBank AJ001712, NCCIA80 (1105).
Encodes a chaperone protein involved in the assembly of mitochondrial complex I (1105).
cit-1 : citrate synthase-1
IIR. Linked to preg (0T/18 asci) (1447).
Cloned and sequenced: Swissprot CISY_NEUCR, EMBL/Genbank M84187, PIR S41563, EMBL NCCIT1A, GenBank NEUCIT1A; EST NM6F5.
Structural gene for citrate synthase, mitochondrial isozyme (EC 4.1.3.7) (627).
cit-2 : citrate synthase-2
VR. Between inl (1T/18 asci) and cya-2 (1T/18 asci) (1447).
Structural gene for citrate synthase, microbody isozyme (EC 4.1.3.7) (627).
cl : clock
VR. Right of pk (1% or 2%) (569, 1965).
Shows noncircadian periodicity. Spreading flat colonial morphology, forming dense bands. The mycelium becomes increasingly dense until growth ceases in all but a few hyphae; these reinitiate the cycle. Grows l cm/day. Band size and period are modifiable. Photographs (569, 2026). Asci from cl ´ wild type are normal. Crosses homozygous for cl give flaccid asci with unordered ascospores, similar to pk ´ pk. So do cl ´ pk, suggesting allelism (1965). pk (bis) sometimes forms growth bands (569). However, substantial crossing-over frequencies and the recovery of pk; cl double mutants (569) indicate that pk and cl are at separate loci. Increased activity of L-glutamine:D-fructose 6-phosphate amidotransferase was found in crude extracts of cl and morphological mutants at five other loci (1758).
cla-1 : clock-affecting-1
Identical to frq7 (397).
cmc : carboxymuconate cyclase
Unmapped.
Cloned and sequenced: Swissprot CCCM_NEUCR, EMBL/Genbank L27538, PIR A55525, GenBank NEUCCMUCY.
Encodes carboxy-cis,cis-muconate cyclase (EC 5.5.1.5) (3-carboxy-cis,cis-muconate lactonizing enzyme) (CMLE) (1306).
cmd : calmodulin
VR. Between inl (3T/18 asci), cya-2 (2T/18 asci) and tom70 (1 or 2T/17 asci). Linked to hsp80, hsp83 (0T/18 asci) (1447).
Cloned and sequenced: Swissprot CALM_NEUCR, EMBL/GenBank L02963, PIR S58703, S28188, S32452, GenBank NEUCLMDLN; EST SP4B1.
Encodes the Ca2+-binding protein that is a key regulator of many cellular processes. Purification (1507) and properties (422). Stimulation of adenylate cyclase (1710); cyclic nucleotide phosphodiesterase (1508, 1898, 2070); protein kinase (2136, 2137). Binds to proteins associated with the cytoskeleton (284, 1506). Neurospora crassa calmodulin activates the calmodulin-dependent protein phosphatase catalytic subunit of calcineurin, which is encoded by cna-1 (874, 1652). It can be ubiquitinated by a Ca2+-dependent mechanism (2303). Calmodulin antagonists affect the circadian rhythm (1432, 2067) and light regulation (1775). Mutants with hypersensitive responses to the calmodulin antagonist chlorpromazine have been isolated [cpz-1 and cpz-2 (2031)]; these mutations affect light regulation and circadian rhythms. Calmodulin antagonists may also affect chitin synthase activity (2024).
cmt : copper-metallothionein
VIL. Between asd-1 (2T/18 asci) and Bml, Cen-VI (3T/18 asci; 1/18) (1388, 1447).
Cloned and sequenced: Swissprot MT_NEUCR, EMBL/GenBank X03009, GenBank NCCMTG, PIR A24641, SMNC; Orbach-Sachs clones X16H10, G4H01, G9G12, G10C10.
Structural gene for copper metallothionein (1387, 1388). RNA level peaks late in the circadian night, but is not induced by light or developmental signals (147). Copper-inducible promoter (1812); metal binding of the polypeptide (151, 602, 1164); purification (1168); use as a heavy-metal bioabsorbent (1538). Strongly resembles a Colletotrichum gloeosporioides polypeptide predicted to be expressed during appressorium formation (943). Allelic with ccg-12.
cna : calcineurin A
VR. Linked near cyh-2 (1652).
Cloned and sequenced: Swissprot P2B_NEUCR, EMBL/GenBank M73032, GenBank NEUCAM.
Encodes calcineurin A, the Ca2+- and calmodulin-regulated protein kinase catalytic (a) subunit of calcineurin (874, 1652). The subunit encoded by cna interacts functionally with the mammalian catalytic subunit (2135). The Neurospora catalytic subunit is partially myristoylated when coexpressed with the mammalian regulatory subunit in insect cells (614). Reduction in cna expression is imposed by antisense RNA expression. Reduction in calcineurin function is imposed by the addition of cyclosporin A or FK506, which increases hyphal branching and changes hyphal morphology, ultimately causing growth arrest (1652). For a review of fungal protein kinases, see ref. (535).
cnb : calcineurin B
Unmapped.
Cloned and sequenced: EMBL/GenBank Y12814, AF034089, GenBank NCCALCINB.
Encodes the regulatory (b) subunit of calcineurin. An insertional mutation at cnb results in swollen and septate hyphae during vegetative growth, indicating that calcineurin may modulate the conidiation pathway (1092).
cni-1 : cyanide insensitive-1
Unmapped. Not in V (592).
Respiration in the first 24 hr of growth is not sensitive to cyanide or antimycin. Initially insensitive to salicylhydroxamic acid (SHAM), but SHAM and cyanide together are inhibitory (592). Cyanide-insensitive respiration and cytochrome c level decrease markedly in late log phase and stationary phase, whereas cytochrome aa3 increases rapidly (1073). By electron microscopy, mitochondria appear defective in early log phase but later resemble wild type (1072). Behaves as a cold-sensitive ribosome assembly mutant. Small subunits are not assembled at low temperatures; the respiratory differences between early and late growth noted previously were found at 30ºC, and normal aa3 production occurred throughout growth at 37ºC (1051). Electron-spin resonance data (591). Selected by failure to reduce tetrazolium in overlay after inositol-death enrichment (593).
cnr : canavanine resistant
IR. Between T(4637)al-1 and In(OY323)R. Therefore, between hom (1%), al-1 and lys-3, nic-1 (11%) (1546, 2064).
Resistance to canavanine (904, 2064, 2065) is due to a constitutive enzyme that cleaves L-canavanine to hydroxyguanidine plus a compound that can be converted to L-homoserine (1218). Resistance, therefore, is dominant. Resistance is altered secondarily by modifiers that affect rate of uptake (1218). Many laboratory strains are resistant, but a few are sensitive (1549). Best scored on 0.2 mg/ml L-canavanine H2SO4, autoclaved in medium, at 2 days and 34ºC (1546, 1585). Sensitive strains have been used to select resistant mutants defective in basic amino acid transport (1736, 1782, 2233). Called can, but pmb rather than cnr is the Neurospora counterpart of CAN in Saccharomyces cerevisiae.
cog : recognition
IR. Between his-3 (1%; 3%) and ad-3A (2%; 7%) (45, 216, 336). Located about 3 kb from the his-3 coding sequence (2275), at the opposite end from the his-3 promoter (216).
The cis-acting recombinator responsible for increasing recombination within his-3 (45) and crossing over between his-3 and ad-3 (336), but only in the absence of rec-2+. Highly polymorphic alleles have been described. cogLa and cogS79a, which are probably identical, mediate high recombination, which is dominant. Low recombination is mediated by cogLA, cogEA, and cogST74A (2276). Generic designations are cog+ for high-recombination alleles and cog for low (2275). The effect of a specific cog allele is limited to the chromatid that contains that allele, as evidenced by crosses heterozygous for translocation TM429, which has a breakpoint between mutant sites within the his-3 locus (336). Recombinators with similar rec specificity are presumably present in other regions under rec-2 control, and cog-like recombinators with differing specificity are presumably located in regions controlled by other rec genes. See rec (Fig. 57).
coil : coiled
IVR. Between arg-2 (2%) and leu-2 (l2%) (141).
Growth of hyphae from ascospores or conidia is initially spiraled in a clockwise coil on the surface of agar or on a membrane placed on agar (Fig. 19). Scorable microscopically in young cultures under low magnification (141). Counterclockwise coiling has been reported for rco-1 (2256).
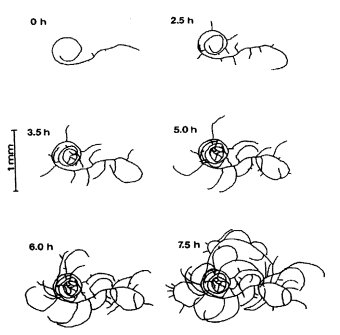
FIGURE 19 A time-lapse sequence of coil-1 hyphae following ascospore germination, on agar medium overlaid with cellophane to prevent hyphae from penetrating the medium. From ref. (141 ).
col : colonial
The name colonial has been used primarily for mutants with restricted mycelial growth that is self-limiting on agar medium. The radial growth of each colony does usually not exceed a few millimeters. Colonial mutants vary widely in texture, density, conidiation, pigment, and fertility. Mutants of the series col-5 to col-l7, described in ref. (719), were in some instances assigned new locus names without having been tested for allelism with already-named morphological mutants having similar map locations. Some were subsequently shown to be recurrences (e.g., col-3 of bn, col-7 of rg-1, col-l4 of sc); others may be recurrences but have never been tested (possible examples are col-5 and col-8 with col-l or cot-1; col-13 and col-15 with vel). Two colonial mutants with nongerminating ascospores are symbolized le-1, le-2. For reviews covering morphological mutants and morphogenesis, see refs. (235), (1281), (1353), (1848), (1851), and (2117). See ref. (384) for growth rates and hyphal diameters of numerous colonial mutants.
col-1 : colonial-1
IVR. Linked to pan-1 (0/47 asci), cot-1 (3%) (102, 708).
No macroconidiation. Growth is cyclic at moderate temperatures and steady at 38ºC. The double mutant col-1; cot-1 grows better than col-1 at 24ºC and better than cot-1 at 38ºC (708). pe; col-1, which forms microconidia (102), was used in early mutation studies, but col-4; pe fl was found to be better (727). The pe; col-1 strain forms microconidia at 25ºC, but macroconidia at 35ºC. Cell-wall analysis; photograph (507). Not tested for allelism with col-5, col-8.
col-2 : colonial-2
VII. Linked to met-7, met-9 (1%), probably to the left (1585, 1592). Right of T(T54M50) (1578).
Colonial morphology (102). Photographs (235, 1851, 1853). Altered NADP-specific glucose-6-phosphate dehydrogenase (EC 1.1.1.49) (242, 1854). (bal and fr are also deficient in G6PD.) Reduced NADPH level (233). Reduced linolenic level (241). Accumulates much neutral lipid (1484). Pyridine nucleotide levels (234). Suppressor su(col-2) increases linear growth rate and influences the electrofocusing pattern of col-2 G6PD (1853). Homozygous col-2 ´ col-2 crosses fail to mature; the immature asci frequently are nonlinear and occasionally show dichotomization (1965). Hyphae swell with age to 20-mm diameter (384).
col-3 : colonial-3
Allelic with bn.
col-4 : colonial-4
IVR. Between met-1 (4%), mus-26 (3%) and arg-2 (1% or 2%) (1369, 1715, 1928).
Spreading colonial morphology, forming dense balls of conidia high in a slant (103). Scorable microscopically after ascospore germination (Fig. 20). Cell-wall-autolyzing enzyme (1247). Reduced amount of cell-wall peptides (2249). The mutant allele is dominant in heterozygous duplications from T(S1229) (110). Used in combination with pe fl to produce microconidiating colonial growth suitable for reversion experiments (727). Called spco-1 (719), c (727).
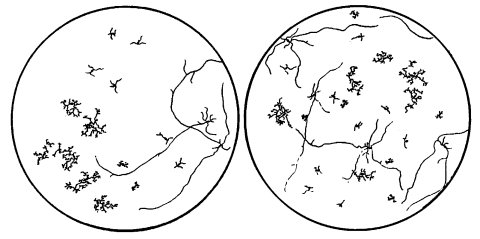
FIGURE 20 A camera lucida drawing of germinating ascospores, showing gene segregation in a cross of col-4 X pyr-2. Ascospores were placed on minimal agar medium in petri dishes, heat-activated at 60C for 30 min, and incubated at 25C for 18 hr. The pyr-2 mutant grows sufficiently on minimal medium to permit the identification of col-4 pyr-2 double mutants. The plate on the left shows segregations in two asci with ascospore pairs isolated in linear order. In the first ascus, genotypes were col+ pyr, col+ pyr, col pyr+, and col pyr+ (from top to bottom). Those in the second ascus were col+ pyr+, col+ pyr, col pyr, col pyr+. The other plate shows the result of plating ascospores from the same cross at random. Genetic analyses can be made directly using either procedure or by transferring germinated ascospores to appropriately supplemented culture tubes for further testing. The random method allows much larger numbers to be observed conveniently. Drawing by M. B. Mitchell. From ref. (2177 ), with permission.
col-5: colonial-5 &# 160;
IVR. Linked to cot-1 (1%), probably to the right (1604).
Dense, nonconidiating, poorly pigmenting, slow-spreading, colonial morphology (719, 1604). Cell-wall analysis and photograph (507). Reverts readily (719). Not tested for allelism with col-l, which it resembles and which maps in the same region. Called col(B28) (1604).
col-6 : colonial-6
IV. Linked to Cen-IV (0/28 asci), pan-1 (21%) (719).
Colonial morphology. Slow ascospore germination (719).
col-7 : colonial-7
Allelic with rg-1.
col-8 : colonial-8
IVR. Linked to pan-1 (4%; 13%) (719).
A fluff of hyphae may form at the top of a slant. Not tested for allelism with col-l or col-5 (719). Peptides in cell wall are reduced in amount (2249).
col-9 : colonial-9
VR. Between inl (16%) and asn (5%) (1383).
Forms small, slow-growing colonies. Reverts readily (719).
col-10 : colonial-l0
IIL. Left of cys-3 (14%), at or near pi. A single wild-type progeny (1/81) from col-10 ´ pi may have been a revertant (719, 1592).
Slow-growing, dense, compact morphology, with no conidia (719). Altered 6-phosphogluconate dehydrogenase (1852, 1854). (col-3 also affects 6PGD). Crosses of col-l0 (R2438) ´ pi (B101) resemble R2438 ´ R2438 and B101 ´ B101 in producing abnormal asci with flaccid walls and unordered ascospores, suggesting allelism (1965). However, R2438 and B101 are distinctly different in morphology. B101 has not been tested for 6PGD. As a marker, pi (B101) is preferable to R2438 because of growth rate, stability, and ease of handling; ro-7 in the same region is preferable to both (1582).
col-12 : colonial-12
I. Linked to mat (l7%; 22%) (719).
Growth from ascospores is slow (719).
col-13 : colonial-13
Probably allelic with vel.
col-14: colonial-14
Allelic with sc (1546).
col-15 : colonial-15
Probably allelic with vel.
col-16 : colonial-16
IIIR. Between acr-2 (6%), pro-1 (10%), ad-4 and leu-1 (1%) (1546).
Balls of powdery conidia are formed at top of slant on glycerol complete medium. Female fertile. A good marker, preferable to com, which grows more slowly and does not conidiate (1582). Complements mo-4 and spco-15 (719).
col-17 : colonial-17
VII. Linked to nt (14%), spco-5 (6%) (719).
Very slow-growing (719).
col-18 : colonial-18
VIR. Between un-23 (3/28) and T(OY320) (1582, 1589).
The original mutant allele was obtained in progeny of Dp(VIR®IIIR)OY329 ´ Normal, presumably as a result of RIP (1589).
col-le : colonial lethal
Changed to le-1.
com : compact
IIIR. Between ace-2 (5%) and ad-4 (<1%; 5%) (1122, 1587).
Forms small, slow-growing colonies (507, 1548). Grows better on complete medium than on minimal medium (1582). Cell-wall analysis. Photograph (507).
Complex I
The NADH:ubiquinone respiratory complex of the inner mitochondrial membrane, with subunits specified both by nuclear genes and by mitochondrial genes. See nuo, ndh. See Appendix 4.
con : conidiation
The symbol con has been used to designate developmentally controlled genes that were identified because their mRNA levels are high during the development of macroconidia (158, 1773, 1956, 1958). con mRNA levels can also increase during microconidia and ascospore formation (1959). The con genes that have been analyzed appear to be subject to complex regulation by environmental and developmental cues; regulatory genes including rco-1, rco-3, wc-1, and wc-2 have roles in their expression. None of the con genes for which null mutants have been obtained appears to be essential for conidiation.
The symbol con was also previously proposed for hypothetical “control of recombination” sites postulated to confer specificity of response to a rec gene product on genes that are subject to rec-mediated recombination control (335). However, no genetic variants are known in the postulated sequences, and there has been no direct demonstration that the sequences conferring specificity of response are in separate cis-acting control genes as distinct from recombinators such as cog. Specificity may be encoded in the recombinator itself. In contrast, con has been widely used as the symbol for genes that are transcribed preferentially during conidiation, and that usage is adopted here.
con-1 : conidiation-1
IIIL. Left of Cen-III, crp-2 (6T/18 asci) (1447).
Encodes conidiation-specific protein 1 (158).
con-2 : conidiation-2
V. Between rDNA (2/18; 4T/18 asci) and con-4a (2/18), lys-1 (8T/18 asci) (1447).
Cloned (158).
Encodes conidiation-specific protein 2 (158).
con-3 : conidiation-3
VI. Linked to Bml (0/18) (1447).
Cloned (158).
Encodes conidiation-specific protein 3 (158).
con-4 : conidiation-4
V. Between rDNA (4/18), con-2 (2/18) and chs-1 (1 or 2/18), am (4/18) (1447).
Cloned (158).
Encodes conidiation-specific protein 4 (158).
con-5 : conidiation-5
IVR. Linked to cot-1 (0/18) (158, 1447).
Cloned (158)
Encodes conidiation-specific protein 5 (158). Induced by blue light (1145).
con-6 : conidiation-6
II. Left of arg-12 (2 or 3/18) (1447).
Cloned and sequenced: Swissprot CON6_NEUCR, EMBL/Genbank L26036, EMBL NCCON6X, GenBank NEUCON6X.
Encodes conidiation-specific protein 6 (158, 2209). The CON-6 protein is present at high levels in free conidia (2209). Putative disruption by RIP caused no obvious conidiation or germination defect (2209). Regulatory studies (84, 1144, 2256).
con-7 : conidiation-7
IIIR. Between ace-2 (4T/18 asci), cyt-8 (3T/18 asci) and ad-2, trp-1 (0/17; 1T/18 asci) (1447).
Cloned (158).
Encodes conidiation-specific protein 7 (158).
con-8 : conidiation-8
IR. Linked to phr (0/18), which maps right of os-1 (15%) in conventional crosses (1908). Linked to nic-1, Fsr-15 (4/18) (1447). The IL location between nit-2 (2/17) and mat (2/17), shown in ref. (1447), appears to be incorrect (1908).
Cloned and sequenced: Swissprot CON8_NEUCR, EMBL/GenBank X07040, GenBank NCCON8, PIR S02210.
Encodes conidiation-specific protein 8 (158, 1731). Regulatory studies (1731).
con-9 : conidiation-9
IVR. Linked to cot-1 (0/18) (158, 1447).
Cloned (158).
Encodes conidiation-specific protein 9 (158), which is expressed at high levels in late-phase vegetative growth and newly formed conidia (1773).
con-10 : conidiation-10
IVR. Linked to cot-1 (0/18) (158, 1447). Adjacent to con-13 (1730).
Cloned and sequenced: Swissprot CONX_NEUCR, EMBL/GenBank M20005, PIR A31849, EMBL NCCON10A, GenBank NEUCON10A. In the same cDNA clone (pCon-10a) with con-13.
Encodes conidiation-specific protein 10 (158, 1730) which has sequence similarities to stress-induced proteins in other organisms (1154). CON-10 polypeptide is localized to the conidiophores where it is seen distributed in the cytoplasm (1957). A RIP mutant in which con-10 and con-13 were disrupted showed no obvious conidiation or germination phenotype (except for not producing these polypeptides) (1957). Fusions of the 5'-region to hph enabled selection for mutants affecting transcript levels of con-10 and other conidiation genes during vegetative growth; mutants isolated in this way include rco-1 and rco-3 (1241, 1242, 2256). Regulatory studies (84, 1144, 1242, 2256). Promoter elements have been defined that are important for regulated expression of con-10 (409, 1153, 1154).
con-11 : conidiation-11
VIR. Right of Cen-VI, Fsr-50 (6T/17 asci). Linked to trp-2 (0/18) (1447).
Cloned (158) and sequenced (184).
Encodes conidiation-specific protein 11 (158), which is induced late in conidiation close to the time of appearance of CON-10 and free conidia. The protein has a predicted size of 51 kDa and an apparent size of approximately 100 kDa. The protein is N-glycosylated and may have other posttranslational modifications. CON-11 can be localized to the periplasmic space by cell fractionation. Normal vegetative transcription is very low, but is slightly derepressed in double-mutant strains with a RIP-inactivated rco-1. Although con-11 is not induced by stress during vegetative growth, the predicted protein sequence is very similar to that of rds1 of S. pombe, a stress-response gene of unknown function (184).
con-13 : conidiation-13
IVR. Adjacent to con-10 (1730), which is linked to cot-1 (158, 1447).
Cloned and sequenced: Swissprot COND_NEUCR, EMBL/GenBank M35120, PIR JH0363, EMBL NCCON13, GenBank NEUCCON13. In the same cDNA clone (pCon-10a) with con-10.
Encodes conidiation-specific protein 13 (814, 1730). CON-13 polypeptide is localized to the conidiophore (1957). A RIP mutant in which con-13 and con-10 were disrupted showed no obvious defects in conidiation or germination phenotype (except for not producing these polypeptides) (1957).
cor-1 : cobalt resistant-1
IIIR. Linked to dow (1%, probably to the right) (2221).
Resistant to inhibition by cobalt (20-fold more resistant than control) and nickel (4-fold more resistant than control), but not by zinc or copper. Cobalt uptake is partially blocked (1374). A cobalt-binding glycoprotein is overproduced (1780).
cot-1 : colonial temperature sensitive-1
IVR. Between pan-1 (2%) and his-4 (1%; 6%) (1369, 1585, 1592).
Cloned and sequenced: Swissprot COT1_NEUCR, EMBL/GenBank X97657, PIR S22711, GenBank NCCOT1.
Encodes a serine/threonine protein kinase (EC 2.7.1.-). The cot-1 gene is homologous to protein kinase genes udk1 of Ustilago, TB3 of Colletotrichum, orb6 of Schizosaccharomyces, the warts tumor suppressor gene of Drosophila, and the myotonic dystrophy kinase gene of humans (570). Two sense transcripts are encoded that differ in length. The deduced transcripts differ in their regulatory domains. The ratio of the transcripts is regulated by blue light and depends on functional products of wc-1 and wc-2 (1142). Null mutations obtained following disruption of the locus by RIP are not remediable, retaining a tight colonial phenotype at all temperatures (2268). The temperature sensitivity reflected in the locus name is due to a posttranslational defect of the original allele, C102t (2268). The following description applies to C102t unless otherwise indicated. Growth is colonial and extremely restricted at 34ºC, with increased hyphal branching, but growth, morphology, and fertility are completely normal at 25ºC and below (Fig. 21). Linear growth is maximum at 24ºC (708), becoming colonial at 32ºC. The mutation is a single-base-pair change in the coding domain. Only one of the two COT-1 isoforms is detectable at restrictive temperatures, and the activity of Ser/Thr kinase is reduced whereas that of types 1 and 2B phosphatase (calcineurin) is increased (760). Colonies from ascospores or conidia are viable at restrictive temperatures and continue to grow slowly with dense branching, but they do not conidiate. They quickly resume normal growth when shifted to a permissive temperature (1369, 2079). Recessive in duplications (1580); apparent dominance in heterokaryons (708) may have resulted from a shift in nuclear ratios. Used in studies of septation and branching (1213), growth-inhibiting mucopolysaccharide (1718, 1719), sulfate transport (1280). Cell wall analysis (708). Growth is stimulated by lysine or arginine (0.1 mM) on glucose medium at high temperatures (1210). Because of its high viability and tightly restricted growth at restrictive temperatures and its normality at 25ºC, cot-1 C102t has had valuable technical applications. For example, crosses homozygous for C102t have been used in combination with sorbose for experiments with rec genes, where high-density ascospore platings are required for precise quantitative analysis of intralocus recombination (332, 1940, 2083). In another application, when C102t is shifted up after initial growth at the permissive low temperature, linear growth is arrested and hyphae assume a “bottle brush” appearance with small side branches (398, 1368) (Fig. 21). This has been used to select ultraviolet-sensitive (UV-sensitive) mutants by subsurface survival on UV-irradiated plates containing p-aminobenzoic acid, which absorbs lethal wavelengths (317, 1839). C102t conidia or ascospores from homozygous C102t crosses have been used for replication in a protocol involving transfer by filter paper (1210). For suppressors of cot-1, see gul, ro. Most suppressors of cot-1 are mutations at various ropy loci, which inactivate cytoplasmic dynein or dynactin (248, 1634).
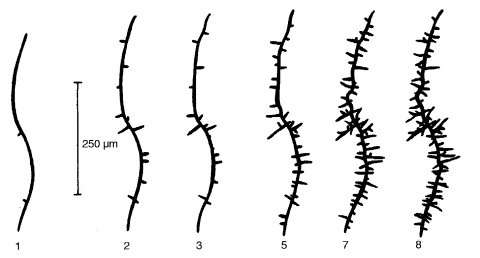
FIGURE 21Branching and arrest of linear growth of cot-1 allele C102 following the transfer of a mycelium from 25 to 37C. Hours following the transfer are given below each drawing. From ref. (398 ), with permission from the British Mycological Society.
cot-2 : colonial temperature sensitive-2
VR. Between pk (8%), ser-2 (5%), T(EB4)L and ad-7 (4%) (321, 1546, 1578, 1603). Recombines with inv (5%) (598).
Colonies are small but fully viable at 34ºC. Growth and morphology are nearly, but not completely, normal at 25ºC (719). Invertase is altered; it is not clear whether cot-2 is the structural gene for a second subunit or whether cot-2 affects invertase structure indirectly, perhaps by altering the carbohydrate moiety (598). See inv. Cell-wall peptides are reduced in amount (2249). Ascospores are round in homozygous cot-2 ´ cot-2 crosses, but are shaped normally in heterozygous crosses (117). Some cot-2 strains carry mei-3, which was discovered in the original cot-2 strain (1469); most strains used by Eggerding et al. (598) are free of mei-3, however, and mei-3 cannot be responsible for the effects on invertase (1476).
cot-3 : colonial temperature sensitive-3
IV. Left of pan-1 (16%; 25%). Probably right of arg-2 (719, 1582).
Forms small, fully viable colonies at 34ºC. Morphology is normal at 25ºC, but the growth rate is reduced (719, 2107, 2109). Septae behind hyphal tips are not plugged (2109).
cot-4 : colonial temperature sensitive-4
VR. Between rol-3 (5%) and inl (10%) (1383).
Forms small colonies at 34ºC. At 25ºC, growth is spreading (719) and the morphology resembles that of cum, sn, and perhaps sp, with late-forming blooms of conidia on aerial hyphae. Female fertility is good in heterozygous crosses, but no perithecia are formed in crosses homozygous for cot-4 (1582).
cot-5 : colonial temperature sensitive-5
IIL. Between T(P2869)R, T(B18)R (1578) and het-c (3%), based on recombination in crosses cot-5 het-C pyr-4 thr-2 ´ het-c and pi het-C pyr-4 thr-2 ´ T(NM149) het-c (924) [these genotypes were given incorrectly or incompletely in ref. (924)].
Little or no growth at 34ºC; colonial at 30ºC. Colonies form short, aerial hyphae at 25ºC (719). Female-sterile. Morphologically distinct from fs-1 (1582).
cox : cytochrome oxidase
Cytochrome c oxidase (EC 1.9.3.1) (shortened to cytochrome oxidase in gene names) is a hetero-oligomeric enzyme located at the inner mitochondrial membrane. Three of its subunits (subunits I, II, and III) are encoded mitochondrially. The remaining subunits are nuclear-encoded.
cox-4 : cytochrome oxidase-4
III. Linked to acr-2 (1 or 2/15) (445), cyt-8 (1766), by RFLP.
Cloned and partially sequenced: Swissprot COX4_NEUCR, EMBL/GenBank M12116, GenBank NEUCOX4.
Encodes cytochrome c oxidase subunit IV (1770). A synthetic peptide corresponding to part of the mitochondrial import-sequence causes increased permeability to small solutes when added to rat liver mitochondria (1942). RFLP data indicate that cox-4 is close to cyt-8; whether they are allelic has not been established.
cox-5 : cytochrome oxidase-5
Allelic with cya-4.
cox-6 : cytochrome oxidase-6
V. Near inl (445).
Cloned and sequenced: Swissprot COX6_NEUCR, EMBL/GenBank M12118, GenBank NEUCOX6.
Encodes cytochrome oxidase subunit VI. Possible allelism with cya-2 not tested.
cox-8 : cytochrome oxidase-8
VIIR. Linked to cit (2T + 1NPD/15 asci), un-2 (2/17), lacc (3/17) (1447).
Cloned (1770).
Cytochrome c oxidase subunit VIII; similar to Saccharomyces cerevisiae cytochrome oxidase subunit VIIa (1767).
cpc-1 : cross pathway control-1
VIL Between ylo-1 (1%; 3%; 143kb), un-13 (105kb) and Cen-VI. Inseparable from T(IBj5)R (457, 1578).
Cloned and sequenced: Swissprot CPC1_NEUCR, EMBL/GenBank J03262, PIR A30208, EMBL NCCPC, GenBank NEUCPC; pSV50 clone 22:11A, Orbach-Sachs clones G2C08, G8C01, G8C03, G15E08, G16H02, G18D02.
Encodes a bZIP transcriptional activator similar to Saccharomyces cerevisiae GCN4 (1525). Simultaneously affects both basal levels and the ability to derepress enzymes in arginine and other amino acid biosynthetic pathways (1768). The CPC-1 polypeptide binds to sequences related to 5'-ATGACTCAT-3' (577); these sequences are present in the 5'-regions of genes controlled by CPC-1. cpc-1 mutations interfere with the cross-pathway activation of amino acid biosynthetic enzymes in response to amino acid starvation or imbalance (1768). cpc-1 mutants are sensitive to 5-MT, 3-AT, FPA, and some nucleoside analogs (326, 1080); also sensitive to high concentrations of some amino acids (121). cpc-1 mutants were originally isolated as arginine auxotrophs by selection in the mutant arg-12s. The cpc-1; arg-12+ single mutant is prototrophic (120, 457). Growth is delayed after ascospore germination; the delay is not alleviated by arginine or precursors. Scorable by delayed growth (120, 457). Sequence changes determined for inactivating mutations CD-15 and IBj-5 (1526). Independently isolated as mts-1 (327, 1080). Independently isolated as lni-1, a regulator of lacc (2043, 2296). cpc-1 mutations are epistatic to the lah-1 mutation, which affects lacc expression (833, 2041), and the lah-1 mutation appears to affect cpc-1 expression (833). T(IBj5) and T(MN9) are inseparable from cpc-1 (1578).
cpc-2 : cross pathway control-2
VIIR. Linked to arg-10 (18%). Right of T(5936) (1099).
Cloned and sequenced: EMBL/GenBank X81875, GenBank NCCPC2, PIR S57839; EST NC4D7.
Encodes a protein entirely composed of WD repeat segments (1386). Simultaneously affects both basal levels and the ability to derepress enzymes in arginine and other amino acid biosynthetic pathways (1386). Sensitive to p-fluorophenylalanine and 3-AT (1386). Female-sterile (1099).
cpc-3 : cross pathway control-3
VR. Linked to cyh-2 (0 or 1/16) (1452, 1795).
Cloned and sequenced: EMBL/Genbank X91867, GenBank NCCPC3.
Encodes the homolog of Saccharomyces cerevisiae Gcn2p. The predicted CPC-3 polypeptide contains an eIF2 kinase domain and a histidyl-tRNA synthetase-like domain. Neurospora strains in which the cpc-3 gene function was disrupted by gene replacement did not activate the transcription of amino acid biosynthetic genes in response to amino acid starvation. Whereas the level of cpc-1 transcript does not appear to be affected in the cpc-3 mutant, the level of CPC-1 polypeptide does not show a large increase in response to amino acid starvation (1795). Thus, cpc-3 has a posttranscriptional effect on cpc-1 expression, presumably at the level of translation, in analogy to the effect of functional GCN2 product on GCN4 translation in Saccharomyces (876, 1768).
cpd-1 : cyclic phosphodiesterase-1
IVR. Right of pyr-2 (19%) (851).
Encodes orthophosphate-regulated cyclic phosphodiesterase (EC 3.4.1.-). Isolated by filtration enrichment from a pho-2 progenitor as cAMP could not be used as the sole phosphorus source (849, 851). Short aerial hyphae on solid medium. Rhythmic conidiation on solid medium resembles that of bd.
cpd-2 : cyclic phosphodiesterase-2
IL. Right of arg-1 (6%) (851).
Impaired activity of orthophosphate-regulated cyclic phosphodiesterase (EC 3.4.1.-). Reduced adenylate cyclase activity. Isolated by filtration enrichment from a pho-2 progenitor as cAMP could not be used as the sole phosphorus source (849, 851). Aerial hyphae are short on solid medium; rhythmic conidiation on solid medium resembles that of bd.
cpd-3 : cyclic phosphodiesterase-3
IL. Right of arg-1 (11%) (852).
Impaired activity of orthophosphate-regulated cyclic phosphodiesterase (EC 3.4.1.-). Reduced adenylate cyclase activity. Isolated by filtration enrichment from a pho-2 progenitor as cAMP could not be used as the sole phosphorus source (852). Hyphal morphology resembles that of cr-1.
cpd-4 : cyclic phosphodiesterase-4
IIR. Right of arg-12 (7%) (852).
Altered activity of orthophosphate-regulated cyclic phosphodiesterase. Reduced adenylate cyclase activity (852). Isolated by filtration enrichment from a pho-2 progenitor as AMP could not be used as the sole phosphorus source. Does not form aerial hyphae or conidia in low-phosphate medium.
cpk : cAMP-dependent protein kinase
IIIR. Linked to un-17 (5%) (1403).
Mutation results in a cAMP-dependent protein kinase activity that is independent of cAMP addition (1403). The function of CPK is unknown at present (1632).
cpl-1 : chloramphenicol sensitive-1
VIL. Between ad-8 (7%; 11%) and aro-6 (5%), lys-5 (6%), un-25 (2%, 4%) (346, 1582, 1815).
The mutant is sensitive to chloramphenicol (<0.5 mg/ml added to autoclaved medium) and antimycin A (1 mg/ml); wild type is resistant to 4 mg/ml of chloramphenicol. Protein synthesis is not grossly altered. Cyanide-insensitive and azide-insensitive respiratory systems are still present. The cytochrome spectrum is normal on minimal medium. Obtained by inositol death enrichment and replica plating, using a strain of genotype inl; trp-3; sn cr-1 (346, 349). Scorable on 0.5 mg/ml chloramphenicol, autoclaved in the medium (1582).
cpt : carpet
IIR. Between arg-5 (3%) and T(NM177)L, pe (6%) (1580, 1585).
Flat, slow-growing mycelium. Aerial hyphae are short, and no conidia are formed unless the culture becomes desiccated (1958). Produces microconidia (793), but much less abundantly than the double mutants fl;dn and pe fl (1582). Homozygous fertile.
cpz-1 : chlorpromazine sensitive-1
Unmapped.
Mycelial growth is hypersensitive to the calmodulin antagonists chlorpromazine, trifluoperazine, imipramine, and W5 (2031). The effects of chlorpromazine on circadian rhythm in wild type are abrogated in the mutant (2031).
cpz-2 : chlorpromazine sensitive-2
Unmapped.
Mycelial growth is hypersensitive to the calmodulin antagonists chlorpromazine, trifluoperazine, imipramine, and W5 (2031). The effects of chlorpromazine on circadian rhythm in wild type are abrogated in the mutant (2031). Growth is temperature-sensitive (1035). Addition of spermidine to the medium or transformation with the spe-3 gene encoding spermidine synthase eliminates the temperature-sensitive phenotype of cpz-2 (1035) and causes circadian rhythm in the mutant to be chlorpromazine- sensitive; growth of mycelia in the mutant remains chlorpromazine-sensitive under these conditions (1035).
cr-1 : crisp-1
IR. Between nic-2 (4%; 7%), ace-7 (1%; 3%) and cys-9 (3%), un-1 (5%) (1414, 1592). Included in duplications from T(4540), which do not include cr-2 or cr-3 (1578). One of the first mutants to be mapped in Neurospora (1200).
Cloned and sequenced: Swissprot CYAA_NEUCR, EMBL/GenBank D00909, GenBank NEUNAC; EST NP2C2; pSV50 clones 1:10G, 4:7C.
Structural gene for adenylate cyclase (EC 4.6.1.1) (2077). The mutant has little or no endogenous adenosine 3¢,5¢-phosphate (cyclic AMP; cAMP) (1524, 2076). Slow mycelial growth. Conidiates rapidly and profusely, close to the surface of agar. Produces very short conidiophores, bearing conidia in tight clusters (1200, 1201). Photographs (1005, 1256). Excellent as a marker. Scorability and viability are good. Morphology may vary on different media. Strains carrying various alleles differ in growth habit. (For example, B123 strains are flat and restricted whereas allele L strains are spreading.) Some cr-1 alleles will conidiate in submerged shake culture (1146, 1633). Mutant B123 can be replicated using a needle replicator (1256). cr-1 ascospores may require longer to mature than cr+. Crosses homozygous for allele B123 exude intact linear asci (1256). Frequently occurring modifier mutations can alter morphology and the ability of cr-1 to use glycerol (720, 1778). For a suppressor of cr-1, see hah.
Used to determine what functions are controlled by cAMP (1524). The abnormal morphology is partially corrected by exogenous cAMP (1742, 1743, 2076, 2077). Guanosine 3¢,5¢-phosphate also stimulates mycelial elongation (1743). Phosphodiesterase inhibitors do not counteract the morphological effect of cr-1 (1743). Other anomalies are also normalized by exogenous cAMP: overproduction and excretion of NAD(P) glycohydrolase (1005); altered induction and localization of b-glucosidase (1779); inability to use glycerol and certain other carbon sources (1147, 2078); limited secretion of extracellular alkaline protease (1798); and ability to use alanine as a gluconeogenic substrate (1460). Cyclic nucleotide levels differ in mycelia and conidia (1742, 1743). The mutant is constitutively thermotolerant (434). Used to study cAMP-binding protein (2104), cell-wall polymer (1916). Called nac (1089). Allele CE4-11-67 called con (1408, 1409).
cr-1 suppresses the apolar growth of mcb at 37ºC. mcb therefore can be used to select new crisp-1 mutations (247). The double mutants sn cr-1 and cr-1 rg-1 form small conidiating colonies suitable for replica plating (349, 1256, 1553, 1830, 1979). These have been used to obtain UV-sensitive mutants (1256, 1830) and to measure recessive lethal mutation (Fig. 22) (1980). The triple mutant sn cr-1; csp-2 can be overlayered without displacing conidia (1454, photograph, 1457).
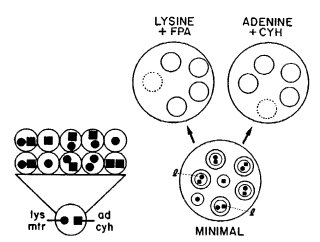
FIGURE 22 The method developed by D. R. Stadler for use of sn; cr-1 strains in quantitative studies of recessive lethal mutation. Mutations are detected using a heterokaryon of constitution (sn cr-1; mtr; lys-5 + sn cr-1 cyh-1; ad-2). ad-2 and lys-5 are complementing recessive auxotrophic markers, and mtr and cyh-1 are recessive mutations conferring resistance to fluorophenylalanine and cycloheximide. sn cr-1 conidia grow into small conidiating colonies suitable for replica plating. (Left) A single conidium containing one nucleus of each genetic type. On minimal medium it grows into a colony that produces many conidia of each of three classes, heterokaryons and the two types of homokaryons. (Right) On minimal medium, the conidia (inner circles) that contain both nuclear types grow into colonies (outer circles) that produce conidia able to grow on replica plates of both test media, unless a recessive lethal mutation has arisen in one of the ancestral nuclei. Reprinted from ref. (1980 ) [Stadler, D., and H. Macleod (1984). A dose-rate effect in UV mutagenesis in Neurospora. Mutat. Res. 127: 39-47.] Copyright 1984, with permission from Elsevier Science.
cr-2 : crisp-2
IR. Between cr-3 (11%), T(NM103) and al-2 (18%); hence, right of thi-1 (18%) (720, 1582).
Conidiation is delayed. Fine, pale-pigmented conidia are then produced in clumps over the agar surface (720). Cell-wall peptides are reduced in amount (2249). Cyclic AMP levels are normal. Overproduces and excretes NAD(P) glycohydrolase; this is not cured by exogenous cAMP (1005).
cr-3 : crisp-3
IR. Between cr-1 (13%), T(4540)R and cr-2 (11%) (720, 1582).
Conidiation is delayed. Fine, pale conidia are then produced uniformly over the agar surface (720). The amount of cell wall peptides is reduced (2249). Cyclic AMP levels are normal. Overproduces and excretes NAD(P) glycohydrolase; this is not cured by exogenous cAMP (1005).
cr-4 : crisp-4
IV. Linked to pdx-1 (6%), cot-1 (22%) (1593)
Resembles cr-1 in appearance (1147).
cr-5 : crisp-5
III. Linked to ad-4 (19%) (974, 975).
Resembles cr-1 in appearance (974, 975).
cre-1 : carbon catabolite regulation
IVR. Right of trp-4 (5 or 6T, 1NPD/17 asci) (1447).
Cloned and sequenced: EMBL/Genbank AF055464.
Homologous to the Aspergillus nidulans carbon catabolite repressor creA and able to bind to CREA target sequences in Aspergillus when expressed heterologously. Used to examine whether the Neurospora gene acts similarly to creA by testing whether CRE-1 and binding sites for CRE-1are important for transcriptional efficiency and glucose regulation of crp-2 and Fsr40 promoters. Disruption of CRE-1 binding had no effect on transcription rate during steady-state growth or carbon shifts, indicating that transcriptional control of ribosomal genes by carbon source is not mediated by cre-1 (480).
crib-1 : cold-sensitive ribosome biosynthesis-1
IV. Linked to met-1 (6%) (1814).
Defective ribosome biosynthesis below 20ºC; attributed to a defect in ribosomal ribonucleic acid (rRNA) processing (1756). Grows at 6% of the wild-type rate at 10ºC and 79% at 25ºC. 37S cytosolic ribosomal subunits are underaccumulated, and relatively little stable 17S rRNA is produced at low temperatures. Not a conditional lethal mutation (1754, 1814). Conditionally defective in the expression of S-adenosylmethionine synthetase activity (1759).
crib(PJ31562) : cold-sensitive ribosome biosynthesis (PJ31562)
IVR. Linked to crib-1 (3%) (1753).
Defective in the biosynthesis of cytosolic ribosomes at 10ºC, but normal at 25ºC. Grows at 16% of the wild-type rate at 10ºC and 90% at 25ºC. Underaccumulates 17S rRNA and, hence, 37S ribosomal subunits. Shows partial complementation in forced heterokaryons with crib-1 (1753).
crp-1 : cytoplasmic ribosomal protein-1
Allelic with cyh-2.
crp-2 : cytoplasmic ribosomal protein-2
III. Linked to Cen-III (0T/18 asci) (1447).
Cloned and sequenced: Swissprot RS14_NEUCR, EMBL/GenBank X53734, GenBank NCCRP2, PIR S11667.
Encodes 40S ribosomal protein S14. Used in studies of promoter elements (435).
crp-3 : cytoplasmic ribosomal protein-3
IVR. Right of trp-4 (3T, 1NPD/18 asci) (1447).
Cloned and sequenced: Swissprot RS17_NEUCR, EMBL/GenBank M63879, PIR S34441, EMBL NCCRP3, GenBank NEUCRP3.
Encodes 40S ribosomal protein S17 (1905).
crp-4 : cytoplasmic ribosomal protein-4
VR. Right of inl (9T/18 asci), tom70 (4T/18 asci), Fsr-20 (1T/18 asci). Linked to sdv-6 (0T/18 asci) (1447).
Cloned and sequenced: EMBL/GenBank AF054907.
Encodes the large subunit of a ribosomal protein homologous to yeast L1 and rat L5, which bind to 5S rRNA (481).
crp-5 : cytoplasmic ribosomal protein-5
Unmapped.
Cloned and sequenced: Swissprot RS26_NEUCR, EMBL/GenBank X55637, PIR S13082, R4NC26, GenBank NEUCRP5A, NCRPSSU, M95597.
Encodes 40S ribosomal protein S26E (2188).
crp-6 : cytoplasmic ribosomal protein-6
Known only as part of the ubi::crp-6 fusion gene, which encodes ribosomal protein S27a.
crp-7 : cytoplasmic ribosomal protein-7
IR. Linked to con-8 (0/18), Tel-IR (1T/18 asci). Adjoins phr (1115a).
Cloned and sequenced: EMBL/GenBank AB015207.
Encodes a ribosomal protein homologous to Saccharomyces cerevisiae YS25 (945).
crp-12 : cytoplasmic ribosomal protein-12
Called rps.
crp(S7) : cytoplasmic ribosomal protein S7
IIIR. Adjoins ro-2 (2154).
Cloned and sequenced: EMBL/GenBank U73847.
Encodes a small-subunit ribosomal protein homologous to the rat ribosomal protein S7 (2154).
csh : cushion
IR. Between thi-1 (12%; 20%) and ad-9 (5%) (1592).
Restricted colonial growth (1585).
csp : conidial separation
In addition to the two genes so named, other mutations are known to result in defective separation of macroconidia. Insertional translocation T(MD2) csp and the duplication progeny it generates are phenotypically similar to csp-1 and csp-2, suggesting that there may be yet another csp locus between arg-5 and pe in IIR (1578). The os-8 mutant and the unmapped histidine kinase nrc genes also are defective in conidial separation.
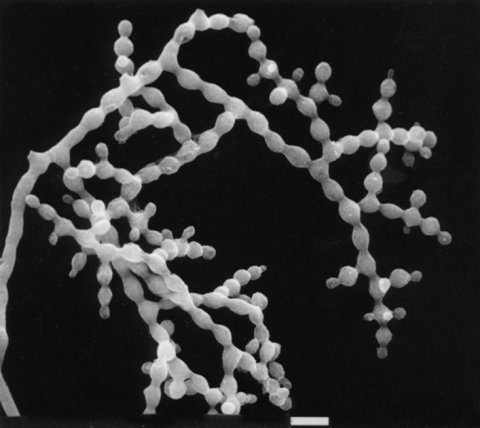
FIGURE 23 The mutant csp-1: conidial separation-1. Bar length-10 mm. Scanning EM photograph from M. L Springer.
csp-1 : conidial separation-1
IL. Between arg-3 (1%) and T(39311)R (1582, 1882).
Conidia fail to separate and become airborne (1882) (Fig. 23). Recessive. Cultures in agar slants are readily scored by a “tap test.” In water, conidia are freed at one-tenth the wild-type concentration (1882). Carotenoids tend to be yellowish in young cultures (1582). Used in connection with bd for the study of circadian rhythms (239). Useful in student laboratories to avoid contamination (1876). Used in combination with csp-2 in a teaching exercise to demonstrate complementation (1880).
csp-2 : conidial separation-2
VIIL. Linked to thi-3 (<1%), probably to the right. Left of T(T54M50), ace-8 (1582, 1882).
Conidia fail to separate and become airborne. Cultures on agar are readily scored by a tap test. Resembles csp-1. Conidia are freed in water suspension, but only long after induction of aerial growth and at 1/100 the concentration obtained using wild type. A csp-1; csp-2 double mutant releases no detectable free conidia under the same conditions (1882). Most csp-2 alleles complement csp-1 in forced heterokaryons to form the wild-type number of free conidia (1882), but csp-2 allele UCLA102 does not (1878). Conidiating colonies of the csp-2; sn cr-1 strain on replica plates can be overlayered without dislodging conidia (1454); photograph(1457).
csr-1 : cyclosporin resistant-1
IR. Right of his-2 (20%) and cyh-1 (1%; 3%) (358, 2111).
Cloned and sequenced: Swissprot CYPH_NEUCR, EMBL/GenBank J03963, X17692, PIR A30809, EMBL NCCYCLO, GenBank NEUCYCLO, NCCYCGEN; EST WO1C2.
Encodes peptidyl-prolyl cis-trans isomerase precursor (EC 5.2.1.8) (PPIase) (cyclophilin) (rotamase) (cyclosporin-A-binding protein) (CPH). Encodes both cytoplasmic and mitochondrial cyclophilins, which differ at their N-termini and which appear to be translated from alternative transcripts differing in their 5'-leader regions (2112). The absence of cyclophilin, or failure to bind it, confers resistance to cyclosporin. Resistance is recessive. Catalyzes protein folding (1826). The mitochondrial enzyme is part of the mitochondrial protein import pathway and interacts with chaperones HSP70 and HSP60 (1702); its function can be discriminated from that of chaperonin (2170). Called cyp.
cum : cumulus
IIIL. Between Tip-IIIL and Sk-2K (2%), Sk-3K (2%), r(Sk-2)-1 (4%), acr-7 (5%; 18%), acr-2 (18%) (280, 1582).
Initially grows as a colony, then spreads and sends up blooms of aerial hyphae that conidiate profusely at the shallow end on agar slants. Good female fertility. Similar in morphology to sn, sp, and cot-4 at 25ºC (1582). Colonies are tighter at higher temperature (1322). Very small inocula (floated conidia) should be used when scoring acr-7 in cum isolates (2121).
cut : cut
IVL. Between cys-10 (28%; 37%) and fi (4%; 10%); therefore, linked to ace-4 (1559, 1582).
The osmotic-sensitive phenotype and the morphology resemble those of mutants named osmotic. Scorable either by failure to grow on medium containing 4% NaCl or by morphology. The morphology approaches normal at high humidity (1116). Conidia do not shake loose when a slant is tapped. The IR linkage shown by the original allele, HK53, was due to an unrecognized I;IV translocation T(HK53), which is inseparable from cut (1580, 1592). An allelic point mutation, LLMI, shows linkage in IVL and segregates independently of I markers (1559). ovc is probably an allele (94).
cwl : cross wall
Defective in septum formation in mycelia and ascogenous cells. Vegetative hyphae tend to bleed, forming an exudate on the agar surface and in lens-shaped pockets beneath. Slow growing, aconidial. Stocks are conveniently kept as heterokaryons with a helper strain such as am1 (1564). The phenotypically normal heterokaryon forms perithecia when it is used as maternal parent. In crosses homozygous for cwl, paraphasoid cells are septate, whereas ascogenous cells contain no cross walls (1680). Helper nuclei from the maternal parent thus contribute to paraphasoidal cells, but not to ascogenous hyphae.
cwl-1 : cross wall-1
IIR. Between arg-5 (3%; 6%) and arg-12 (12%) (1582).
Hyphal septa are largely absent. Subject to alteration by modifiers that restore septa and increase the growth rate, but the original mutant gene can be extracted by crossing. Recessive (719, 825, 1582). Called mo(R2441).
cwl-2 : cross wall-2
IIR. Between arg-12 (10%), un-20 (4%) and fl (9%) (1546).
Hyphal septa are largely absent. Recessive in heterokaryons, but the cross-wall phenotype appears erratically. Mutant allele 1098 originated in flm-1 (1546).
cy : curly
IL. Linked to arg-1 (0/34), ad-5 (1/54), probably to the left (1582).
Young hyphae are seen to be curled when examined under low magnification on agar or on the walls of culture tubes. Growth rate and gross morphology are normal (1366). Conidia fail to shake loose in a tap test, as with csp mutants (1955). Called c.
cya-1 : cytochrome a-1
IL. Linked to mat (6%, shown to the left) (170).
Deficient in cytochrome aa3. Cannot reduce tetrazolium. Very slow growth. Female sterile (170). Possibly not really a cya mutant (162).
cya-2 : cytochrome a-2
VR. Linked to al-3 (3%, indicated to the right) (170). Linked to inl (one apparent NPD, 0T/18 asci), cmd, hsp80, hsp83 (2T/18 asci) (1447).
Deficient in cytochrome aa3. Cannot reduce tetrazolium. Slow growth. Female-sterile (170). Possible allelism with cox-6 not tested.
cya-3 : cytochrome a-3
VIL. Between chol-2 (10%) and cyt-2 (10%) (170).
Deficient in cytochrome aa3. Cannot reduce tetrazolium. Slow growth (170). The aa3. deficiency is suppressed by antimycin A (167). Spectrum (167).
cya-4 : cytochrome a-4
IIR. Between arg-5 (2/39; 1T/18 asci) and preg (1T + one double exchange/18 asci). Linked to Fsr-55 (0T/18 asci) (1339, 1447, 1769).
Cloned and sequenced: Swissprot COX5_NEUCR, EMBL/GenBank E91991, M12117, M24389, PIR A30134, B25629, OTNCV, GenBank NEUCOX5, NEUCOXV; Orbach-Sachs clone X13G05, G22A10.
Structural gene for cytochrome c oxidase subunit V (1769). Deficient in cytochrome aa3. Cannot reduce tetrazolium. Growth is slow. Spectrum (167, 170). Called cox-5.
cya-5 : cytochrome a-5
IVR. Right of pan-1 (2%) (1437).
Cloned and sequenced: EMBL/GenBank AF002169, EMBL NCAF2169.
Homologous to yeast PET309, which regulates the expression of mitochondrially encoded COXI (388). Deficient in cytochrome aa3. Subunit 1 polypeptide of cytochrome c oxidase is absent by immunological criteria and by analysis of mitochondrial translation products. Growth is slow. Recovery from crosses is poor (10%). Selected as a tetrazolium nonreducer (173, 1437, 1438). The circadian period is shortened (236, 560). Called cya-U-34.
cya-6 : cytochrome a-6
IVR. Right of pan-1 (2%) (1437).
Deficient in cytochrome aa3. Alleles called cya-6-2 and cya-6-35 are heat-sensitive. Selected at 41ºC by slow growth on salicylhydroxamic acid and resistance to tetrazolium. At least five subunits of cytochrome c oxidase are present at 41ºC by immunological criteria, but they are not associated. Complements cya-5 (1437).
cya-7 : cytochrome a-7
VR. Right of Cen-V. An earlier report of linkage in III was incorrect (162).
Deficient in cytochrome aa3. Allele cya-713 is heat-sensitive. Selected at 41ºC by slow growth on salicylhydroxamic acid and resistance to tetrazolium. At least five subunits of cytochrome c oxidase are present at 41ºC by immunological criteria, but they are not associated (1437). Called cya-7-13.
cya-8 : cytochrome a-8
VIIL. Between Tip-VIIL and T(ALS179) (5%), cyt-7 (7%), adh (19%), nic-3 (39%) (1593)
Deficient in cytochrome aa3; phenotypically similar to [mi-3] (162). Very slow, sparse, transparent growth with no or little conidiation. Severity of the defect is variable with mutants of independent origin. Many germinants from ascospores fail to survive, but they can be rescued readily in a heterokaryon with aml ad-3B cyh-1. New cya-8 mutations are found in ~20 % of the progeny of eas(UCLA191) (1546).
cya-9 : cytochrome a-9
IVR. Left of trp-4 (9%) (162).
Deficient in cytochrome aa3. Mitochondrial-encoded subunit 1 of aa3 is of a larger apparent size than in wild type; mitochondrial-encoded subunit 3 is less abundant than in wild type (173).
cyb-1 : cytochrome b-1
VR. Between al-3 (24%) and his-6 (10%) (170).
Deficient in cytochrome b. Cannot reduce tetrazolium. Slow growth. Spectrum (167, 170). Suppresses the aa3 deficiency of cyt-2 and the mitochondrial mutant [mi-3] (167).
cyb-2 : cytochrome b-2
Unmapped. A report of VI linkage (170) may be incorrect (162).
Deficient in cytochrome b; erratic deficiency of aa3 (162). Cannot reduce tetrazolium. Very slow growth. Reduced female fertility. Spectrum (167, 170). Shortened circadian period (238, 244, 560).
cyb-3 : cytochrome b-3
IIL. Between Tip-IIL and ro-3 (9%) (1546).
Deficient in cytochrome b. Cannot reduce tetrazolium. Heat-sensitive. Grows slowly, with mutant phenotype at 38 or 39ºC, but nearly normal mycelial growth rate at 25 and 34ºC (2207). Growth from germinated ascospores is delayed at 34ºC (1546). The circadian period is shortened (560).
cyc-1 : cytochrome c-1
IIR. Between thr-3 (35%; 38%) and trp-3 (15%; 18%) (170, 2015).
Cloned and sequenced: Swissprot CYC_NEUCR, EMBL/GenBank X05506, NCCYTCR, NCCYC1R, PIR A00041, CCNC.
Structural gene for cytochrome c. Growth rate is reduced; female sterile (170). Mutations called cyc-1-1 and cyt-12 are allelic (0/1000 recombination; no complementation) (201). Strains carrying the allele called cyt-12 (or cyc-1-12 or cyt-12-1) are partially deficient for cytochrome aa3, whereas cyc-1-1 is not. The cytochrome aa3 level in mitochondria is temperature-sensitive (2015). Import of apocytochrome c into mitochondria is blocked by an mRNA splicing deficiency in cyc-1-1 (201). Although the gene was named cyt-12 when first mapped, it has been called cyc-1 in recent publications (201, 2015). cyc-1 is the preferred name because it is more explicit and more accurate.
cyh-1 : cycloheximide resistant-1
IR. Between nit-1 (6%) and T(STL76), al-2 (8%; 13%) (929, 1554, 1580).
Resistant to cycloheximide (929, 1458). Resistance is recessive in duplications (2122). Dominance reported in forced heterokaryons (929, 1458) may have been due to skewed nuclear ratios (2122). Protein synthesis on ribosomes of the mutant cyh-1 proceeds in the presence of cycloheximide in a cell-free system (1643). Readily scored on slants with 10 mg/ml cycloheximide autoclaved in the medium. Excellent as a marker and valuable for selecting somatic recombinants or deletions in heterozygous duplications (1458, 2123). Used as a component of heterokaryons for testing spontaneous mutations in mutator strains (1010). Used to show that the cycloheximide-induced phase shift of the circadian clock involves protein synthesis (1435). Cycloheximide induces changes in the pattern of synthesized polypeptides in wild type, but not in the mutant (1609). Called act-1: actidione resistant-1.
cyh-2 : cycloheximide resistant-2
VR. Between lys-2 (<1%) and leu-5 (1% or 2%), sp (2%; 9%) (929, 1582, 1603).
Cloned and sequenced: Swissprot RL2A_NEUCR, EMBL/GenBank X06320, PIR S01744, R6NC7A, EMBL NCCRP1G, GenBank NCCR; Orbach-Sachs clones X23C08, G16H11.
Encodes 60S ribosomal protein L27A, homologous to yeast CYH2 (1096). Resistant to cycloheximide (929, 1458). Protein synthesis on mutant ribosomes proceeds in the presence of cycloheximide in a cell-free system (1643). Excellent as a marker, readily scored on slants with 10 mg/ml of cycloheximide autoclaved in the medium or with 1 mg/ml added after autoclaving. Resistance in heterokaryons has been reported to be dominant (929, 1234) or recessive (1840); dominance may depend on nuclear ratios or on the medium. A modifier of resistance has been isolated (1745). Used in mutagenicity test systems (1234). Used to show that the cycloheximide-induced phase shift of the circadian clock involves protein synthesis (1435). The double mutant cyh-1; cyh-2 grows slowly and is much more insensitive to cycloheximide than is either single mutant (929). Called act-1, crp-1.
cyh-3 : cycloheximide resistant-3
Unmapped. Unlinked to cyh-2. Stated to be distinct from cyh-1 (2169).
Resistant to 100 mg/ml of cycloheximide. The double mutant cyh-2; cyh-3 is morphologically abnormal, resistant to more than 2400 mg/ml (2169). The one known allele, CH96, was called act-5 (2168), act-3 (2169).
cyp : cyclophilin
See csr-1.
cys-1 : cysteine-1
VIL. Between cys-2 (1%; 3%) and ylo-1 (8%) (1414, 1986).
Uses sulfite, thiosulfate, cysteine, or methionine. The original isolate (allele 84605) also had a partial requirement for tyrosine and showed high tyrosinase activity at 25ºC, but not at 35ºC (903, 1414, 1901). These properties reverted, whereas the cysteine requirement is stable (903). Used in studies of intra- and interlocus recombination (1414, 1987, 1989, 1994-1996).
FIGURE 24 A speculative model of the sulfur regulatory circuit of N. crassa. The positive- acting cys-3 gene is itself expressed only when the cells become limited for sulfur. CYS-3 is a trans-acting DNA-binding protein that shows autogenous control, increasing its own expression, and that also turns on cys-14, ars, and other structural genes of the circuit. CYS-3-binding sites have been identified in the 5'-promoter regions of ars, cys-14, cys-3, and scon-2. The sulfur-controlled genes scon-1 and scon-2 prevent CYS-3 function during conditions of sulfur catabolite repression. The scon-1 product is postulated to be the factor that recognizes the metabolic repressor, cysteine. The SCON-2 -transducin protein alone does not inhibit cys-3 function, but apparently requires modification by or interaction with the unknown product of scon-1. An active form of SCON-2 or a SCON-1-SCON-2 complex is postulated to inhibit the transcription of cys-3 directly (as diagrammed) and/or by binding to the CYS-3 protein to prevent its positive autogenous control. Note that CYS-3 positively controls scon-2, which negatively controls cys-3, thus constituting a complex set of feedback regulatory loops. Interactions that have been demonstrated experimentally are shown with solid lines, whereas speculative interactions are shown with dashed lines. Solid circles represent known CYS-3-binding sites, and open circles represent presumed CYS-3 binding sites. The gene specifying choline sulfatase has not been identified. From ref. (1287 ), with permission from the Annual Review of Microbiology, Volume 51, 1997, by Annual Reviews (www.annualreviews.org).
cys-2 : cysteine-2
VIL. Between un-4 (4%), T(T39M777), T(Y18329) and cys-1 (1%; 3%) (1414, 1578, 1986).
Uses cysteine or methionine. Strains carrying cys-2 alleles are heterogeneous in response to thiosulfate, but do not use sulfite (1414). Lacks sulfite reductase, as do cys-4 and cys-10 (1160) (Fig. 45). No interallelic complementation. Used in studies of intra- and interlocus recombination (see cys-1).
cys-3 : cysteine-3
IIL. Between ro-7, pi (4%) and T(AR18)L, pyr-4 (18%; 21%) (1414, 1580, 1592).
Cloned and sequenced: Swissprot CYS3_NEUCR, EMBL/Genbank M26008, PIR A30225, A37066, EMBL NCCYS3, GenBank NEUCYS3.
Encodes a nucleus-localized, bZIP DNA-binding protein that regulates genes of sulfur uptake and metabolism (e.g., sulfate permeases, aryl sulfatase, choline sulfatase) (539, 1279, 1287, 1329, 1519) (Fig. 24). Uses sulfite, cysteine, or methionine; little or no response to thiosulfate (1279, 1414). Grows well on methionine. Resistant to chromate (1279). Consensus DNA-binding site (1191). Studied extensively (1281). Functional analysis (414, 686, 689, 1518). Conserved in Neurospora intermedia and in Neurospora crassa Oak Ridge and Mauriceville wild types (1287). Expression from a heterologous promoter in N. crassa causes the expression of sulfur-controlled genes (1517). Expression positively autoregulated; also regulated by sulfur catabolite repression and by scon-1 and scon-2 (1287). Mutation of the F-box of the regulator scon-2 eliminates cys-3 expression (1111). Transcriptional and posttranscriptional regulation of cys-3 RNA levels (1287). Regulation of CYS-3 polypeptide half-life by sulfur (2054). cys-3 ascospores darken slowly or not at all, even when a cys-3 strain is the fertilizing parent and when a strain carrying the heat-sensitive allele NM27t is crossed at 25ºC, the permissive temperature for growth. cys-3 can be used effectively as an autonomous ascospore color mutant for demonstrating segregation patterns in asci (1584) (Fig. 25). Adding of methionine to crossing medium promotes darkening but fails to give good recovery of cys-3 progeny. Recovery of a few percent cys-3 progeny is possible in well-aged crosses (1414, 1582).

FIGURE 25 Maturing asci from a cross heterozygous for cys-3, which has pleiotropic effects on ascospore maturation and cysteine biosynthesis. Mature asci show four black and four white spores. The white ascospores received the cys-3 mutation. Asci with all eight spores unpigmented are immature. One ascus at top center and two asci at upper left show first-division segregation. The remaining mature asci show second-division segregation patterns resulting from crossing over between cys-3 and the centromere. The length of mature ascospores is ~26 mm. Photograph by N. B. Raju. From ref. (1676 ), with permission from Urban & Fischer Verlag.
cys-4 : cysteine-4
IVR. Between rug (10%), T(NM152)R and uvs-2 (5%) (1414, 1578, 1580, 1591, 1993).
Uses cysteine; slight response to thiosulfate (1414). Poor growth on methionine. Lacks sulfite reductase, as do cys-2 and cys-10 (1160) (Fig. 45).
cys-5 : cysteine-5
IL. Between leu-4 and ser-3 (0.1%) (1592, 2198). Linked to cys-11 (<1%) but probably at a distinct locus, with the order leu-3 cys-5 cys-11 mat in a cross showing no negative interference (1415).
Uses sulfite, thiosulfate, cysteine, or methionine (1414). Lacks 3'-phosphoadenosine-5'-phosphosulfate reductase (EC 1.8.99.4) [F.-J. Leinweber, cited in refs. (1414) and (1415)] (Fig. 45). Enzymatically distinct from cys-11 (adenosine 5'-triphosphate sulfurylase), which it complements in heterokaryons [F.-J. Leinweber, cited in refs. (1414) and (1415)]. Leaky, but not so as to interfere with scoring. Ascospores may be oozed from perithecial beaks rather than being forcibly expelled. For good recovery of cys-5 progeny, crossing media should be supplemented even when the protoperithecial parent is cys+; otherwise cys-5 ascospores may fail to blacken. cys(NM86) and cys(85518), which initially were listed as cys-5 alleles, are now designated cys-11.
cys-6, -7, -8: cysteine-6, -7, -8
Lost. Identity with other loci was never established (1414, 1619).
cys-9 : cysteine-9
IR. Between cr-1 (3%) and T(4540)R, thi-1 (13%) (1414, 1578).
Cloned and sequenced: Swissprot TRXB_NEUCR, EMBL/Genbank D45049, EMBL NCCYS9, GenBank NEUCYS9.
Structural gene for thioredoxin reductase (EC 1.6.4.5) (1499). Circadian rhythm is affected in the mutant (1499). Uses sulfite, thiosulfate, cysteine, or methionine. Somewhat leaky (1414)
cys-10 : cysteine-10
IVL. Between Tip-IVL, T(AR33) and acon-3 (1%; 6%), ace-4 (19%; 33%), cut (28%; 37%). Linked to uvs-3 (3%; 7%) (1122, 1414, 1578, 1596).
Uses cysteine, cystathionine, homocysteine, or methionine, with a slight response to thiosulfate (893, 1160, 1414). Lacks sulfite reductase, as do cys-2 and cys-4 mutants (1160) (Fig. 45). Käfer (1008) has obtained good growth on thiosulfate. Growth is better on casein hydrolysate than on methionine (1546). cys-10 chol-1 double mutants grow better on methionine alone than on methionine plus choline (1414). Name changed from met-4; see ref. (1414).
cys-11 : cysteine-11
IL. Linked to cys-5 (<1%). Between leu-3 (8%) and mat (5%). Probably a locus distinct from cys-5, with the order leu-3 cys-5 cys-11 mat in crosses showing no negative interference (1414, 1415).
Uses sulfite, thiosulfate, cysteine, or methionine (1414). Affects adenosine 5'-triphosphate sulfurylase (EC 2.7.7.4) [1278; F.-J. Leinweber, cited in refs. (1414) and (1415)] (Fig. 45). Enzymatically distinct from cys-5 (3'-phosphoadenosine-5'-phosphosulfate reductase), which it complements in heterokaryons [F.-J. Leinweber, cited in refs. (1414) and (1415)]. Mutant cys(85518) also lacks adenosine 5'-triphosphate sulfurylase (1278) and thus is allelic with cys-11 rather than with cys-5, in harmony with recombination data (1415). Called cys(NM86) (1414).
cys-12 : cysteine-12
IR. Linked to al-1 or al-2 (0/76). Right of ad-9 (12%) (1415).
Uses cysteine or methionine (1415). No information on what precursors are used.
cys-13 : cysteine-13
IR. Right of his-3 (2%), tyr-2, T(T112M4i) ad-3B (1279, 1546, 1578).
Resistant to chromate. No demonstrable requirement. Deficient in sulfur permease I (conidial type) (1278, 1279) (Fig. 24). Scored on minimal agar medium with 25 mM chromate and 0.25 mM methionine after 3 days or longer at 34°C. (cys-3 mutants are also chromate-resistant.)
cys-14 : cysteine-14
IV. Linked to cot-1 (21%) (1279).
Cloned and sequenced: Swissprot CY14_NEUCR, EMBL/Genbank M59167, PIR A37956, GenBank NEUSPII; pSV50 clone 16:2B.
Structural gene for sulfate permease II (mycelial type) (1046, 1278, 1279) (Fig. 24). CYS-14 is a membrane protein with 12 predicted membrane-spanning domains (1046) and was the first member of an evolutionarily conserved transporter protein superfamily to be found (1287, 1783). Localized to the plasma membrane (984). CYS-3 regulatory elements in the promoter have been defined (1193). mRNA half-life measured (1046). The single mutant is deficient in sulfate transport in the mycelial stage, but is sensitive to chromate. Resistant to selenate. The double mutant cys-14; cys-13 cannot transport inorganic sulfate, but grows on methionine; both single mutants are prototrophic (1279).
cys-15 : cysteine-15
IVR. Between pdx-1 (0T/55 asci), T(S1229)L and arg-14, met-1 (3%) (111, 1490, 1580).
Uses sulfite, thiosulfate, cysteine, or methionine, but is unable to use sulfate (1414, 1490). Only one allele is known, with the requirement not separated from a deficiency of D-amino acid oxidase (1490); a closely linked coincident lesion is thought to be responsible. (See oxD for D-amino acid oxidase mutants that have no cysteine requirement.) Called cys(oxD1).
cys(oxD1):
Changed to cys-15.
Cytochrome
Cytochrome-deficient Mendelian mutations have been subdivided into four main classes: cyt, deficiency of more than one cytochrome; cya, deficiency of cytochrome aa3; cyb, deficiency of cytochrome b; and cyc, deficiency of cytochrome c (170, 1631). Apart from diagnostic spectra, they are characterized by slow growth (4-7 days for conidiation of mutants vs 3 days for the wild type) and by the inability to reduce tetrazolium (170). These properties have been used in screening new mutants. cni also affects cytochrome spectra (592). The relation of tet (tetrazolium-resistant nonreducer) to cytochrome mutations is not known. Cytochrome defects that result from mutations of the mitochondrial genome (e.g., [mi-1], [mi-3]) are not considered here except as they interact with chromosomal genes such as su([mi-1]); see Appendix 4.
cyt-1 : cytochrome-1
IL. Between leu-3 (5%; 8%) (1128) and T(OY321), leu-4 (1599).
Cloned and sequenced: Swissprot CY1_NEUCR, EMBL/Genbank X05235, NCCYTC1R, PIR A27187.
Structural gene for cytochrome c (202). Deficient in cytochromes aa3 and b. Very slow growth. Female sterile (170, 1371). Growth is slower on organic complete medium than on minimal, presumably owing to inhibition by yeast extract (1371, 1592). Mutant cyt(U-9) is linked and may be an allele of C115, the original cyt-1 mutation (162, 170).
cyt-2 : cytochrome-2
VIL. Between cya-3 (10%) and lys-5 (6%) (170, 1371, 1986)
Cloned and sequenced: Swissprot CCHL_NEUCR, EMBL/Genbank J05075, PIR A34365, EMBL NCCYC8, GenBank NEUCYC8.
Structural gene for cytochrome c heme lyase (EC 4.4.1.17) (551). Completely deficient in cytochromes aa3 and c. Very slow growth. Female sterile. Complements cyb-2 (170, 1371). Spectrum (170). Cytochrome aa3 deficiency is suppressed by cyb-1 and by antimycin A (167).
cyt-3 : cytochrome-3
Allelic with cyt-4.
cyt-4 : cytochrome-4
IR. Between the breakpoints of T(AR173); hence, between Cen-I, os-4, sn, and nuc-1, lys-4 (1578).
Cloned and sequenced: Swissprot CYT4_NEUCR, EMBL/Genbank M80735, PIR A38227, GenBank NEUPROPHOS.
Encodes a mitochondrial protein homologous to cell-cycle protein phosphatase (2115). The mutant is deficient in mitochondrial RNA processing (544) and hence, deficient in cytochromes aa3 and b. Grows slowly. The circadian period is altered (560). The phenotype is partially suppressed by the electron transport inhibitor antimycin (166). The cyt-4 -7 mutant and alleles 5, 10, and 14 listed next are all defective in the splicing of large mitochondrial rRNA (162, 166). cyt-4 complements mutants at cyt-18 and cyt-19, genes that are also required for the splicing of large mitochondrial rRNA (166). Five alleles are cold-sensitive. Mutants called cyt-3-5, cyt-U-10, and cyt-U-14 (170) proved to be cyt-4 alleles and were designated cyt-4-5, cyt-4-10, and cyt-4-14, respectively (162).
cyt-5 : cytochrome-5
IVR. Left of trp-4 (9%). Linked to cyt-19 (<1%; 5%; 10%) (170). Near sdv-11, sdv-12 (162).
Cloned and sequenced: Swissprot RPOM_NEUCR, EMBL/Genbank L25087, EMBL NCCYT5MIT, GenBank NEUCYT5MIT.
Deficient in cytochromes aa3 and b. Slow growth. Not defective in the splicing of mitochondrial rRNA, unlike cyt-19, which is closely linked but complements cyt-5 (166, 170). A clone that includes the sequence for mitochondrial RNA polymerase rescues the mutant phenotype (369). Allele cyt-5-4 was called cyt-4.
cyt-6 : cytochrome-6
VII. Linked to wc-1 (2%). Indicated to the left (170).
Deficient in cytochromes aa3 and b. Grows slowly. Not tested for allelism with slo-2 (170).
cyt-7 : cytochrome-7
VIIL. Between T(ALS179) and adh (9%), nic-3 (18%) (1546).
Deficient in cytochromes aa3 and b. Slow growth. Possibly allelic with su([mi-1])-5 (170). Allele 20 is cold-sensitive (1546).
cyt-8 : cytochrome-8
IIIR. Linked near ad-4 (170). Between ace-2 (1 or 2T/18 asci) and ad-2 (4T/18 asci), trp-1 (3/18) (1447).
Deficient in cytochromes aa3 and b. Scored by slow growth on glycerol at 15 or 18ºC. Possibly allelic with su([mi-1])-1 (170). RFLP data indicate cyt-8 is close to cox-4; whether they are allelic has not been established.
cyt-9 : cytochrome-9
VL. Between lys-1 (5%) and T(MB67)R, Cen-V (4T/57asci), at (2T/57 asci; 5%) (170, 1602).
Deficient in cytochromes aa3 and b. Slow growth (170).
cyt-12 : cytochrome-12
Changed to cyc-1.
cyt-15 : cytochrome-15
VIR. Between pan-2 (4%) and trp-2 (3%) [H. Bertrand, unpublished, cited in ref. (2151)].
Mitochondria of the mutant are deficient in cytochromes aa3 and b, whereas cytochrome c is present in higher than normal amounts. Involved in the assembly of small subunits of mitochondrial ribosomes [H. Bertrand, unpublished, cited in ref. (2151)].
cyt-18 : cytochrome-18
IR. Linked to al-2 (10%) nic-1 (1%; 5%) (399, 1262).
Cloned and sequenced: Swissprot SYYM_NEUCR, EMBL/GenBank M17118, PIR A27158, SYNCYT, GenBank NEUTYRSM.
Encodes tyrosyl-tRNA synthetase (EC 6.1.1.1) (26). Heat-sensitive; grows slowly. Stimulated by the addition of tyrosine. Deficient in cytochromes aa3 and b at 37ºC (1631). Mitochondrial protein synthesis and the assembly of small mitochondrial subunits also are abnormal (401). A novel large RNA precursor (35S) is found at restrictive temperatures (401). The intervening sequence of large (25S) mitochondrial rRNA is not excised. Used for direct measurement of tyrosyl-tRNA synthetase participation in RNA splicing (26, 285). Alleles 289-67 and 299-9 differ in the speed of turn-off of RNA processing when the temperature is shifted (1262). Complements cyt-4 and cyt-19, which are also required for splicing large mitochondrial rRNA (166).
cyt-19 : cytochrome-19
IVR. Between T(S4342)L, arg-14 and T(NM152) L, pyr-3 (5%). Linked to cyt-5 (<1%; 5%; 10%) (166, 170, 1377).
Deficient in cytochromes aa3 and b. Slow growth. Required for splicing of mitochondrial large rRNA. Mutant is defective in intron splicing from mitochondrial transcripts (402). Complements cyt-5. Complements cyt-4 and cyt-18, which are also required for splicing (166). Called cyt-U-19 (170).
cyt-20 : cytochrome-20
IL. Linked to mat (0/151) (399).
Cloned and sequenced: Swissprot SYV_NEUCR, EMBL/Genbank M64703, PIR A41251, GenBank NEUXXX.
Encodes cytoplasmic and mitochondrial valyl-tRNA synthetase; possibly has other functions (1104). Deficient in the small subunit of mitochondrial ribosomes, but contains normal ratios of 19S to 25S rRNA in whole mitochondria (401). Either the cyt-10 mutation is allelic with un-3, or, if un-3 strain 55701 is complex, cyt-10 is a component. The cyt-10 component is responsible for the temperature sensitivity of strain 55702, which is also ethionine-resistant. The transport deficiency responsible for ethionine resistance is not temperature-sensitive. cyt-20 has also been called cyt(289-56) (1104). See un-3.
cyt-21 : cytochrome-21
IL. Between ro-10 (1%) and nit-2 (5/18), Fsr-12 (9T/18 asci). Linked to mus-40, pzl-1 (1/18) (976, 1447). An earlier report of linkage in II (399, 1596) was incorrect.
Cloned and sequenced: Swissprot RT24_NEUCR, EMBL/Genbank J03533, X06360, PIR A29927, EMBL NCCYT21, GenBank NEUCYT21.
Encodes mitochondrial ribosomal protein S-24 (1107). Allele 297.24 (1631), called cyt-21-1, inactivates an RNA splice site; activation of a cryptic splice site causes an in-frame deletion in the polypeptide (1108). At 25ºC, deficient in cytochromes aa3 and b. Defective in mitochondrial ribosome assembly. Deficient in the small subunit of mitochondrial ribosomes. The 19S rRNA is degraded rapidly. The effect is more severe at higher temperatures. No growth at 40ºC. The phenotype resembles that of the mitochondrial mutant [mi-1] (401), but the deficiency is not suppressed by su([mi-1])-4 or su([mi-1])-5 (399). Called cyt(297.24).
cyt-22 : cytochrome-22
IIIL. Linked to cum (6%), acr-7 (19%) (1134). Left of con-1 (4/15), cyt-8 (5/18) (1447).
Encodes a mitochondrial ribosomal protein. Also called cyt(289-4) (1103)
cyt-25 : cytochrome-25
VI? Linked to pan-2 (32%) (162)?. Does not complement cyt-15 in heterokaryons (2151).
Cytochromes aa3 and b are deficient, whereas cytochrome c is present in higher than normal amounts. Deficient in large subunits of mitochondrial ribosomes, whereas small subunits appear to be in excess. Female-sterile with no functional protoperithecia, but fertile as male parent (2151). Called cyt-U-28.
cyt(289-56)
Changed to cyt-20.
cyt(297-24)
Changed to cyt-21.
cyt-U-28: cytochrome-U-28
Changed to cyt-25.
Return to the 2000 Neurospora compendium main page