e : heterokaryon incompatible-e
Used in early publications as a symbol for het-e.
eas : easily wettable
IIR. Between ace-1 (1%) and fl (9%) (1546).
Cloned and sequenced: Swissprot RODL_NEUCR, EMBL/GenBank X62170, X67339, PIR A46222, GenBank NCBLI7DNA.
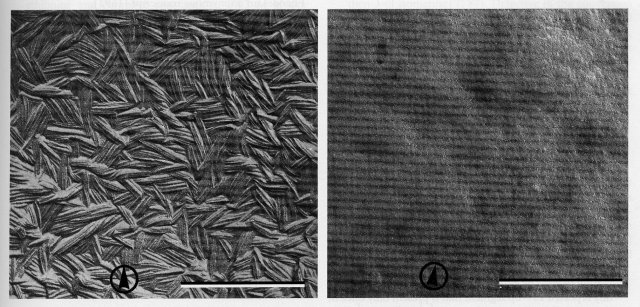
Specifies a self-assembling protein that forms a hydrophobic layer of rodlets on the surface of conidia. This remarkable rodlet hydrophobin is soluble only in 100% trifluoroacetic acid (2071). In the eas mutant, conidia are devoid of rodlets (142) and, hence, easily wettable (1875) in contrast to the hydrophobic wild type (Fig. 26). eas superficially resembles csp mutants in that conidia do not readily become airborne, but it differs from csp in that conidia do not remain joined in the proconidial chains (1875, 1958). It is somewhat sensitive to high osmotic pressure, and aging cultures may appear brighter orange than wild type. Conidiating cultures are readily scored by tapping an inverted slant, transferring conidia to water, or adding a drop of water to the culture. Transcription of eas is inducible by blue light and is under circadian control (145, 586, 1143, 1227), peaking late at night (147). Clock, light, and developmental regulation are controlled by distinct cis-acting upstream elements (145). Induction of eas by light does not require an intact circadian oscillator (60). Transcription of eas is normal in the mutants acon-2 and acon-3, but not in fl (60). eas allele UCLA191 is inseparable from a rearrangement that blocks recombination distal to the eas locus; this led originally to incorrect mapping of eas at the IIR tip. Allele KH5.9 is a translocation (1578). Location of eas right of ace-1 is based on crosses using RIP-derived point mutant JD105 (1546). Because conidia do not become airborne, double mutants have been constructed that combine eas with markers used for teaching (700, 1790). Genes originally identified as clock-controlled gene ccg-2 and blue-light-inducible gene bli-7 proved to be eas alleles (145, 586, 1019, 1143).

FIGURE 26 Replicas of the surface of macroconidia of wild-type (left) and the mutant easily wettable (right), showing the absence of rodlets in eas. Bar length = 0.5 mm. Scanning EM photograph. Reprinted with permission from Nature [ref. (142 )]. Copyright 1978 Macmillan Magazines Ltd.
eat : encodes anonymous transcript
Six genes closely flanking the mating-type idiomorphs were defined on the basis of transcripts, with the order eat-6 eat-5 mat eat 1 eat-2 eat-3 eat-4. Those right of mat are located in the polymorphic region called “variable” in ref. (1690). Regulation differences and the phenotype of a mutant inactivated by RIP indicate that genes in this region are concerned with mating and sexual development. Transcripts of eat-1 and eat-2 are unique in showing mating-type-specific differences in size and expression (1691).
eat-1 : encodes anonymous transcript-1
IL. Right of mat. Between mat and eat-2 (1691).
Cloned and sequenced: EMBL/GenBank AF037226.
Transcript size and expression are different for the two mating types. Attempts to identify alleles inactivated by RIP have been unsuccessful. The alleles are perhaps pseudogenes, reflecting the accumulation of recessive mutations resulting from close linkage to mat (1691). For other genes expressed differentially in the two mating types, see ccg-4, eat-2, mfa-1, and scp.
eat-2 : encodes anonymous transcript-2
IL. Right of mat. Between eat-1 and eat-3 (1691).
Cloned and partially sequenced: EMBL/GenBank AF037227.
Transcript size and expression are different for the two mating types. The eat-2 allele in mat A contains a tandemly repeated 119-bp segment that is present only once in the eat-2 allele of mat a. The sequenced region is >95% similar in mat A and mat a. A portion of a 909-bp ORF corresponding to the first 58 amino acids of the putative product shows 95% identity to a domain present in pma-1 (plasma membrane ATPase-1). [This homology was shown first by S. M. Mandala (1925)]. A mutant eat-2 allele, obtained by RIP in a mat A strain, is abnormal in both vegetative and sexual phases. Growth is very slow but is better on crossing medium (with no ammonium nitrogen) than on Vogel’s minimal growth medium. No conidia are formed. Perithecia are unbeaked and devoid of ascospores when the mutant is crossed as either male or female. The mutant vegetative phenotype is recessive. The mutant sexual phenotype appears to be dominant. Alternatively, normal fertility may be dependent on the presence of functional eat-2 sequences from both mating types. A few ascospores can be obtained from crosses when the male parent carries the mutant eat-2 allele in a heterokaryon with the am1ad-3B cyh-1 inactive-mating-type helper strain. These ascospores can germinate, but the germlings are inviable (1691). For other genes expressed differentially in the two mating types, see ccg-4, eat-1, mfa-1, and scp.
eat-3 : encodes anonymous transcript-3
IL. Right of mat. Between eat-2 and eat-4 (1691).
Cloned.
Encodes as anonymous transcript (1691).
eat-4 : encodes anonymous transcript-4
IL. Right of mat. Between eat-3 and the right end of the variable flanking region (1691).
Cloned.
Encodes an anonymous transcript (1691).
eat-5 : encodes anonymous transcript-5
IL. Left of mat. Between eat-6 and mat (1691).
Cloned.
Encodes an anonymous transcript (1691).
eat-6 : encodes anonymous transcript-6
IL. Left of mat. Between un-3 and eat-5 (1691).
Cloned.
Encodes an anonymous transcript (1691).
edr-1 : edeine-resistant-1
VI. Linked to ad-1, pan-2 (<1%) (2069).
Resistant to 200 mg/ml edeine. Only a fraction of edr conidia grow. Recessive. Called edr-1 (2069).
edr-2 : edeine-resistant-2
VIL. Left of ad-1 (19%) (2069).
Resistant to 200 mg/ml edeine. Only a fraction of edr conidia grow. Recessive (2069). In intact cells, edeine inhibits the synthesis of protein, DNA, and RNA of the wild type but not of the mutant. In vitro, edeine inhibits protein synthesis equally for both mutant and wild type. Hence, uptake is probably impaired in the mutant (2172). Used to examine the effect of edeine on crossing over and intragenic recombination (2068). Called edr-2, edr-29.
eif5A : eukaryotic initiation factor 5A
Unmapped.
Cloned and sequenced: Swissprot IF5A_NEUCR, EMBL/GenBank U02638, PIR S55278, EMBL NCIF5A, Genbank NCEIF5A.
Encodes eukaryotic initiation factor 5A (eIF-5A homolog) (2050), which is a substrate for deoxyhypusine synthase, the product of dys-1 (2050, 2051).
en(am)-1 : enhancer-1 of am
VR. Between am (8%) and inl (1%). Linked to gln (1%) (256, 642, 647).
In en(am)-1 am double mutants, en(am)-1 blocks the adaptation of am on minimal medium that is devoid of amino nitrogen (647) (Fig. 49). The double mutants are inhibited by ammonium and grow adequately only when glutamate is the sole nitrogen source. The en(am)-1 single mutant grows well on minimal medium, but is unable to use, as the sole nitrogen source, proline (256), methionine, alanine, isoleucine, valine, urocanate, hypoxanthine, uridine, urea, or bovine serum albumin (350). Relative resistance to p-fluorophenylalanine or ethionine and complete resistance to 0.02 M glycine cosegregate with en(am)-1 (642). Glutamate synthase (GOGAT) is normal (566). The single mutant is scored using proline as the sole N source (256) or (better) by resistance to 0.2 mM p-fluorophenylalanine (642). Called I (566).
en(am)-2 : enhancer-2 of am
IIR. Linked near pe (1906).
en(am)-2 counteracts the leakiness of am on minimal medium. The en(am)-2; am double mutant grows well on L-alanine (25 mM), 0.5% casein hydrolysate (1906), or glutamate (5 mM) (566). The en(am)-2 single mutant grows normally on minimal medium. en(am)-2 lacks glutamate synthase (GOGAT) (Fig. 50). The double mutant en(am)-2; am lacks both NADP-glutamate dehydrogenase (1738) and glutamate synthase activities (566). Frequent revertants of en(am)-2; am on suboptimal medium are attributed to back-mutation at am (1906). Called en-am.
En(pdx) : Enhancer of pdx-1 pigment
IL. Linked to mat (5%), probably to the left (1274).
The En(pdx); pdx-1 double mutant excretes yellow pigment when grown on Vogel's medium (2163) or Westergaard and Mitchell's medium (2208) supplemented with suboptimal pyridoxine plus ammonium sulfate at 5 mg/ml, but not at 1 mg/ml. Pigment is not produced by either single mutant (1274). The addition of a nonlimiting concentration of pyridoxine inhibits production of the pigment on both media. Production of the pigment also is inhibited in heterokaryons between complementing pdx alleles. En is dominant over En+ in heterokaryons between noncomplementing pdx alleles (1662).
er : erupt
Allelic with rg-1.
erg : ergosterol
Ergosterol mutants have been detected by resistance to nystatin and other polyene antibiotics. The known mutants are female-sterile. Intercrosses for allelism tests can be made, however, by using a heterokaryon as the female parent (787, 1385). Biosynthesis is illustrated in Fig. 27.
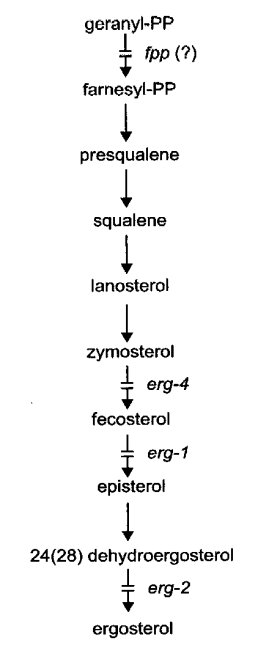
FIGURE 27 The probable pathway of sterol biosynthesis, showing sites of gene action (787 , 1385 ). From ref. (1596 ), with permission from the American Society for Microbiology.
erg-1 : ergosterol-1
VR. Between pk (2%) and asn (9%) (787).
Cloned and sequenced: EMBL/GenBank U59671, Swissprot ERG2_NEUCR; Orbach-Sachs clone G17F12.
Lacks fecosterol D8, D7-isomerase [5-ergosta-8,24(241)-dien-3-ol isomerase] (787). Defective in the conversion of fecosterol [5-ergosta-8,24(241)-dien-3-ol] to episterol (Fig. 27). Ergosterol is deficient in the cell membrane, conferring strong resistance to nystatin and other polyene antibiotics (785, 787). Infertile as female. Slow growth, reduced conidiation. The nysr mutants of ref. (1385) are blocked in the same reaction, but have not been tested for allelism with erg-1.
erg-2 : ergosterol-2
VR. Left of inl (6%) (787).
Ergosterol is deficient in the cell membrane, thereby conferring slight resistance to nystatin and other polyene antibiotics (784, 785). Lacks 24(28)-dehydroergosterol hydrogenase or reductase (EC 1.-.-.-), which mediates the terminal step of ergosterol synthesis (787) (Fig. 27). Poorly fertile as female. Good growth and conidiation.
erg-3 : ergosterol-3
IIIR. Between T(AR17)R, dow (10%, 14%) and Tip-IIIR (1582).
Cloned and sequenced: Swissprot ERG3_NEUCR, EMBL/GenBank X77955, GenBank NCERG3, PIR S44170; Orbach-Sachs clone G18A10.
Encodes sterol Δ14,15-reductase (EC 1.-.-.-) (1533). Ergosterol is deficient in the cell membrane. Resistance to nystatin and other polyene antibiotics is increased slightly (784, 785). Female-sterile, but tiny protoperithecia are formed. Growth is slow, conidiation is reduced, and the production of aerial hyphae is uneven. Mutants are resistant to the steroidal glycoside a-tomatine and sensitive to the phytoalexins pisatin and biochanin A (1532, 1894). The erg-3 null mutant is complemented by sequences encoding the transmembrane domain of human lamin B receptor, indicating defective sterol C-14 reductase activity (1647).
erg-4 : ergosterol-4
IR. Linked to al-1 (10%) (1582).
Ergosterol is deficient in the cell membrane, conferring a slight resistance to nystatin. Lacks C-24 (zymosterol)-methyl transferase (EC 2.1.1.41). Accumulates zymosterol (Fig. 27). Infertile as female (787). Growth is slow; colonial at 34ºC, spreading at 25ºC (1582).
erp38 : ERP38 protein
Unmapped.
Cloned and sequenced: Swissprot ER38_NEUCR, EMBL/Genbank Y07562, GenBank NCERP38.
Encodes a putative protein disulfide isomerase ERP38 precursor (EC 5.3.4.1), a stress-inducible member of the PDI family (986).
esr : enhancer of spermidine requirement
Mutations that increase the need for spermidine. The trait is scored by combining the putative esr mutation with a leaky spe mutation and testing strains on polyamine inhibitors dicyclohexylamine or a-difluoromethylornithine (DFMO). Because arginine imposes a spermidine requirement on strains carrying aga, these can also be used. Ascospores and (usually) conidia of esr mutants germinate and then pause, often for more than 1 day, before continuing to grow. The known esr mutations, originally called BMH (464), were named and given locus numbers by ref. (451).
esr-1 : enhancer of spermidine requirement-1
VR. Between pyr-6 and inl (7%) (464).
Increases the need for spermidine (464).
esr-2 : enhancer of spermidine requirement-2
I. Linked to mat (10%) (464).
Increases the need for spermidine (464). A similar mutant, esr(BMH8), is linked to mat at 3% and may be allelic.
esr-3 : enhancer of spermidine requirement-3
Unmapped. Not linked to mat (1L) or inl (VR) (464).
Increases the need for spermidine (464).
eth-1 : ethionine resistant-1
IL. Between arg-1 (<1%) and arg-3 (<1%) (1336, 1592).
Cloned and sequenced: Swissprot METK_NEUCR, EMBL/GenBank U21547, PIR S65800.
Structural gene for S-adenosylmethionine synthetase (EC 2.5.1.6) (981, 1045, 1298) (Fig. 45). Heat-sensitive, not growing at 37ºC (1336). Resistant to ethionine at 24ºC (1336). Resistance attributed to the overproduction of methionine (1028, 1030) and/or reduced production of the toxic S-adenosylethionine (98, 981). In eth-1+/eth-1 partial diploids, ethionine resistance is dominant but temperature sensitivity is recessive (98). Deficient in DNA methylation at semipermissive growth temperatures (660). Levels of several enzymes that are normally repressible by methionine are not repressed by methionine in eth-1r even at growth-permissive temperatures (260, 1324, 1874). Both heat sensitivity and ethionine resistance are reparable by high osmotic pressure (1324). For high-resolution analysis of this chromosome region, see refs. (1299) and (1740).
eth-2 : ethionine resistant-2
Provisional name for a mutational lesion present, together with cyt-20, in the un-3 mutant 55701. See un-3.
exo-1 : exoamylase-1
IL. Linked to mat (7%), ad-3A (22%). Stated to be left of mat (615).
Shows hyperproduction of b-amylase, a-amylase, glucoamylase, invertase (b-fructofuranosidase), pectinases, and (to a lesser extent) trehalase (432, 433, 764, 765, 1641, 1954, 2002). Enzymes are secreted abundantly upon depletion of an exogenous carbon source (765). A polysaccharide is also released (615). Conidial enzyme levels are increased sevenfold. Amino sugar content of the cell wall is altered (764, 765). The initial allele was called SF26, exoa-1 (615, 765). A probable second allele was found in inl 89601 a (372, 615, 2002). With allele SF26, high amylase and high invertase cosegregated in 91 isolates (764). With alleles from the 89601 strain in a mixed-background cross, high amylase and high invertase each acted as if due to a single major gene with many modifiers; high amylase and high invertase usually cosegregated and were not correlated with alkaline phosphatase levels (2002). The relation of exo-1 to gene VI-178, which reverses the repression of invertase and trehalase production by mannose (1323), is not known. For a linked gene defective in glycoamylase, see sor-4, the T9 allele of which was called gla in ref. (106) and amy in ref. (615). exo-1 was not tested for allelism with sor-4 T9, but is stated to map on the opposite side of mat. Because inl 89601 a has been used to obtain mutants by inositol-less death enrichment, exo-1 may be present but unrecognized in some laboratory stocks.
Return to the 2000 Neurospora compendium main page