f : fast
Changed to su([mi-1])-f.
fas : fatty acid synthesis
Changed to cel.
fba : fructose bisphosphate aldolase
Unmapped.
Cloned and sequenced: Swissprot ALF_NEUCR, EMBL/GenBank L42380, Genbank NEUF16B; EST NC4F12.
Encodes fructose 1,6-bisphosphate aldolase (EC 4.1.2.13) (2259).
fdh : formate dehydrogenase
VR. Contiguous with leu-5 (380).
Cloned and sequenced: Swissprot FDH_NEUCR, EMBL/GenBank L13964, PIR A47117, EMBL NCFDHA, Genbank NEUFDHA; EST NM1D6.
Structural gene for formate dehydrogenase (EC 1.2.1.2). Under developmental control. Expressed during conidiation and early germination (380).
fdu-1 : fluorodeoxyuridine resistant-1
Allelic with ud-1.
fdu-2 : fluorodeoxyuridine resistant-2
IVR. Right of cys-4 (2%) (265).
Resistant to 5-fluorodeoxyuridine, 5-fluorouracil, and 5-fluorouridine. Resistance is partially dominant in heterokaryons. Involved in the regulation of pyr-3, udk, and ud-1 (267). Scored by spotting a conidial suspension on medium containing 4 ´ 10-5 M 5-fluorodeoxyuridine, filter-sterilized (885).
fes-1 : iron-sulfur subunit-1
Unmapped.
Cloned and sequenced: Swissprot UCRI_NEUCR,EMBL/Genbank X02472, GenBank NCCYRFES.
Encodes the iron-sulfur subunit of ubiquinol-cytochrome c reductase (EC 1.10.2.2) (841).
ff : female fertility
Infertile as female, but fully fertile as male (fertilizing) parent. Besides those listed, 32 more ff mutants were obtained by (1000). These are not listed separately because they are unmapped and lack locus numbers. These comprise 28 complementation groups. They have been characterized with respect to the position of the block in perithecial development, dominance, effects on vegetative growth, supplementability, and independence of mating type (1000). These mutants are available from FGSC. A different symbol, fs (female sterile), has been used by other workers for mutants having the same phenotype. See fs for a listing of additional mutants, including some that have other names and that show impaired female fertility as part of a pleiotropic syndrome.
ff-1 : female fertility-1
IIR. Between aro-1 (5%) and un-20 (4%) (2044).
Female sterile, with no protoperithecia (2044, 2045). Glycerol utilization is enhanced. The carbon source affects vegetative expression (2098). Aconidial on liquid glycerol medium. Conidiation is good, but aerial hyphae are reduced and surface growth is heavy on sucrose minimal slants. Growth on glycerol reduces levels of both pyruvate dehydrogenase and dihydrolipoyl transacetylase (418). Conveniently scored by failure to produce protoperithecia on small slants of synthetic cross medium (7 days, 25ºC). Allele T30 was originally recognized as female-sterile and called ff-1 (2044, 2045). Allele JC744 was first characterized by glycerol utilization and called glp-3 (418).
ff-2 : female fertility-2
Unmapped.
No protoperithecia. Normal vegetative morphology (880).
ff-3 : female fertility-3
IR. Right of os-1 (3%) (368).
Defective in protoperithecial production. Abnormal morphology. Found in strain T22, which also contains ty-3 and ty-4. Not allelic to ty-3 or T (368, 881). Not tested for allelism with so.
ff-5 : female fertility-5
IIIR. Between pro-1 (2%) and met-8 (1%) (2044).
Produces sterile brown protoperithecia without trichogynes. Darkens medium. Vegetatively normal. Closely resembles ff-6. Not allelic with ty-1 or ty-2 (2044).
ff-6 : female fertility-6
IIIR. Linked near ty-1 (880).
Produces many large black protoperithecia, but no perithecia are formed after fertilization. Black pigment is excreted into the medium (880).
fi : fissure
IVL. Between ace-4 (10%), cut (4%: 10%) and pyr-1 (10%; 15%), pdx-1 (31%; 44%) (1122, 1582).
Produces exudate in fissures formed under the agar and on the surface. Variable expression. Best scored on minimal synthetic cross medium (2208) at 34ºC, pH 6. Some isolates difficult to score (1122, 1585).
fkr-1 : FK506 resistant-1
VR. Between leu-5 (4%) and ure-2 (13%) (119).
Resistant to inhibition by the immunosuppressant FK506 (123). Phenotypically normal in other respects, including sensitivity to cyclosporin A. Immunodetectable FK506-binding protein is present in the resistant mutant (123). Incompletely dominant to wild type in heterokaryons.
fkr-2 : FK506 resistant-2
VR. Between inl (5%) and cot-2 (3%) (123).
Cloned and sequenced: Swissprot FKBP_NEUCR, EMBL/GenBank X55743, PIR S11090, GenBank NSFKBPA; EST NC4A7.
Resistant to inhibition by the immunosuppressant FK506. Phenotypically normal in other respects, including sensitivity to cyclosporin A. Lacks immunodetectable FK506-binding protein (FKBP), which functions as a peptidyl-prolyl cis-trans isomerase (EC 5.2.1.8). Incompletely dominant to wild type in heterokaryons (123).
fl : fluffy
IIR. Between ace-1, eas (9%) and mus-23 (1%, 5%), trp-3 (3%) (85, 1592). In a common cosmid with mus-23 (85).
Cloned and sequenced: EMBL/GenBank AF022648; Orbach-Sachs clones X24A11, G15G05.
Encodes a developmentally regulated transcription factor (C6 zinc cluster protein) required for normal conidial morphogenesis, resulting in mycelia devoid of macroconidia (Fig. 28). Mutant alleles have been sequenced (85). Development of macroconidia is blocked in null mutants (1199, 1958). Because fluffy strains are aconidiate and highly fertile (1202), they are used routinely as female parents in tests for mating type and for chromosome rearrangements (1558, 1595, 1606). Produces few microconidia when dry, but when wetted produces sufficient microconidia to have been used in early irradiation and mutation studies (1204, 1787). Large numbers of microconidia can be obtained under certain conditions (1744, 2120, 2219). Double mutants pe fl (102, 1389) and fl; dn (1563) produce abundant microconidia; fl; dn (but not pe fl) is fertile when homozygous. Photograph of microconidial formation (1510); see also ref. (1744). SEM photograph (1958). Nuclear numbers in microconidia (102, 133, 902). Mutant allele flY, which has highly conservative missense mutations (85), is exceptional in producing macroconidia, and these are yellow (1594). Cell-wall analysis (403). Immunoelectrophoretic pattern (1539). Paradoxical high alcoholic glycolysis on nitrate medium (159). Deficiency of isocitrate lyase on acetate medium (2117). Used to study the regulation of al-1, al-2, and al-3 (1188). When fl strains of opposite mating type are inoculated separately on crossing medium in plates, a double line of perithecia forms where they meet, similar to the barrage in Podospora (776, 780) (Fig. 37). fl ascospores show high spontaneous germination (1546, 1673, 2201). Allele C-1835 was called acon (1409, 1585).

FIGURE 28 The mutant fl: fluffy. Bar length = 10 mm. Scanning EM photograph from M. L. Springer.
fld : fluffyoid
IVR. Left of his-5 (2%) (1928). Near arg-14 (2255).
Resembles fl in appearance, usually forming no macroconidia (1585) or only minor constrictions (1958). Capable of producing conidia when dessicated or starved, however (573) (Fig. 29).
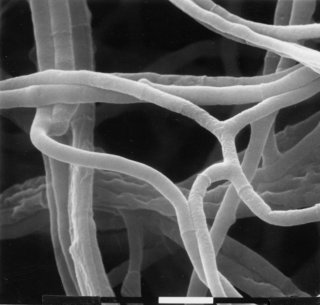
FIGURE 29 The mutant fld: fluffyoid. Bar length = 10 mm. Scanning EM photograph from M. L. Springer.
flm-1 : flame-1
An os-1 allele.
flm-2 : flame-2
An os-4 allele.
fls : fluffyish
IR. Between nit-1 (5%; 19%) and al-1 (6%; 19%) [P. St. Lawrence, cited in refs. (1546) and (1548)].
Cultures are initially aconidiate, resembling fl, but are capable of conidiating later. Vegetative growth is slow (1546, 1958). Shows a suboptimal growth response to methionine (1548). Called un(STL6).
fmf-1 : female and male fertility-1
I. Between mat (2%; 15%) and cr-1 (2%). Linked to arg-1 (1003).
When fmf-1 is present in either parent, male or female, perithecial development is blocked 15 hr after fertilization, prior to meiosis. Perithecia attain only 40% normal diameter. Recessive in heterokaryons. Can be crossed as one component of a heterokaryon, either as female or as male. Female fertility is also restored in mixed-mating-type (fmf-1 A + fmf+ a) heterokaryons that are homokaryotic for tol (1003). Called PBJ6 (1002).
for : formate
VIIR. Between wc-1 (6%) and frq (2%), oli (5% or 6%), dr (3%), un-10 (6%) (1094, 1228, 1585, 1604)
Cloned and sequenced: Swissprot GLYC_NEUCR, EMBL/GenBank, M81918, PIR A42241, Genbank NEUSERHMT; EST NC5A6; pSV50 clone 31:5E.
Structural gene for cytosolic (but not mitochondrial) serine hydroxymethyltransferase (EC 2.1.2.1) (260, 412, 1308) (Fig. 45). Expression is subject to cross-pathway control by cpc-1 (1308). The mutant grows well on formate (0.3 mg/ml, autoclavable) or formaldehyde and has increased formyl THF synthetase, methylene THF dehydrogenase, isocitrate lyase, and glyoxalate aminotransferase activities (411, 412). Growth is aided slightly if glycine, histidine, or choline is added to formate (843). The for mutant also responds to supplements that either spare or supply methylene tetrahydrofolate. Will grow on a mixture of methionine and adenine and suboptimally on adenine alone (843). In auxanograms, for responds weakly to adenine or ascorbate alone, but strongly to adenine plus either histidine, tryptophan, or ascorbate [for possible rationale, see ref. (1472)]. Serine hydroxymethyltransferase purification (1100). Used to examine folate storage in dormant conidia (1101) and pathways of one-carbon metabolism (989). Folic acid auxotrophs have not been found in Neurospora, presumably because folate is not transported into the cell.
fox-2 : fatty acid oxidation-2
Unmapped.
Cloned and sequenced: EMBL/GenBank X80052, NCFOX2, PIR S54786; EST NM4H8.
Structural gene for a multifunctional b-oxidation protein involved in fatty acid degradation (664).
fpp : farnesyl pyrophosphate synthetase
Unmapped.
Cloned and sequenced: Swissprot FPPS_NEUCR, EMBL/GenBank X96944, GenBank NCFPPSGEN, PIR S71434, S71436.
Encodes farnesyl pyrophosphate synthetase (EC 2.5.1.10). Involved in membrane sterol synthesis. Not induced by light (891).
fpr : fluorophenylalanine resistant
The mapped fpr mutants were selected as FPA-resistant. Many mutants isolated by resistance to FPA were found to be bradytrophs (very leaky auxotrophs); most of these required one of many amino acids. The resistance, which disappears when the required growth factor is added, may arise from trans-inhibition of amino acid transport or by induction of the cross-pathway control response (1059, 1768). It is not established whether the mapped fpr mutants are bradytrophs.
fpr-1 : fluorophenylalanine resistant-1
VR. Linked to cyh-2 (<1%) (1067, 2230).
Resistant to p-fluorophenylalanine and 4-methyltryptophan. Isolated as resistant to FPA in the presence of su(mtr)-1. Resistance is recessive in a heterokaryon. Used in a mutagenicity test system (1234). Suppressed by several lys and arg auxotrophs, which apparently give the double mutant greater sensitivity to FPA with no increase in uptake (1067). Scored on solid medium containing 10 mg/ml FPA or 60 mg/ml MT (1057).
fpr-3 : fluorophenylalanine resistant-3
IIIR. Linked to trp-1 (<1%), thi-2 (5%) (1057).
Resistant to FPA but not to 4-methyltryptophan. Not resistant in the presence of indole. Amino acid uptake is normal through both transport systems I and II, as defined in ref. (1057). Isolated in su(mtr). Scored on solid medium containing 10 mg/ml FPA (1057), added before autoclaving.
fpr-4 : fluorophenylalanine resistant-4
VR. Right of inl (11%) (1057).
Resistant to p-fluorophenylalanine and 4-methyltryptophan. Isolated in su(mtr). Not tested for amino acid uptake. Scored on solid medium containing 10 mg/ml FPA or 60 mg/ml MT (1057).
fpr-5 : fluorophenylalanine resistant-5
I. Left of al-2 (25%) (1057).
Resistant to p-fluorophenylalanine but not to 4-methyltryptophan. Isolated in wild type. Not tested for amino acid uptake. Scored on solid medium containing 10 mg/ml FPA (1057).
fpr-6 : fluorophenylalanine resistant-6
VIR. Between pan-2 and trp-2 (477).
Resistant to p-fluorophenylalanine. The only allele, UM300, was found as a variant unable to take up arginine to satisfy the requirement of arg mutants. This blockage is manifest mainly when ammonium is in the medium. Uptake of many other metabolites (amino acids, uridine, sugars) is also affected. The primary defect is unknown (451, 477). FPA resistance is suppressed by am (1335). Not tested for allelism with cpc-1 or mod-5, which map in the same area and cause increased rather than decreased uptake. Called UM300, fpr(UM300).
fr : frost
IL. Between ro-10 (18%) and T(OY330)L, un-5 (6%) (1578, 1582).
Cloned and sequenced: EMBL/GenBank AB021703, Orbach-Sachs clone X18G08 (1946a).
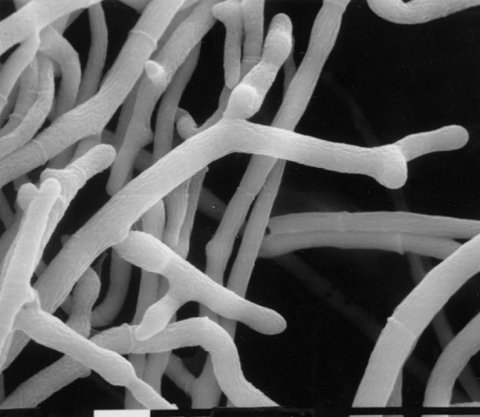
Delicate multiple hyphal branching on an agar surface (719) (Fig. 30). Aerial growth is coral-like, with no blastoconidia (1548, 1955) (Fig. 31). Deficient in G6PD (EC 1.1.1.49) (as are col-2 and bal) (1854, 1857). Addition of Ca2+ almost completely corrects the abnormal branching (533). Partially deficient in linolenic acid (241). Morphology is partially corrected by exogenous linolenic acid (1743, 1847). Cell-wall peptides are reduced in amount (2249). Low adenylate cyclase activity and low cyclic AMP (1847, 1855). Used to determine what functions are controlled by cAMP (1524). Unlike cr-1, the morphology of fr is not corrected by exogenous cyclic nucleotides (1743, 1856). Morphology was reported to be corrected by theophylline (1847), but in another study (1743) no correction by phosphodiesterase inhibitors was seen. Cell-wall analysis; photograph (235, 507, 1851). Recessive in duplications (1580). Female sterile.
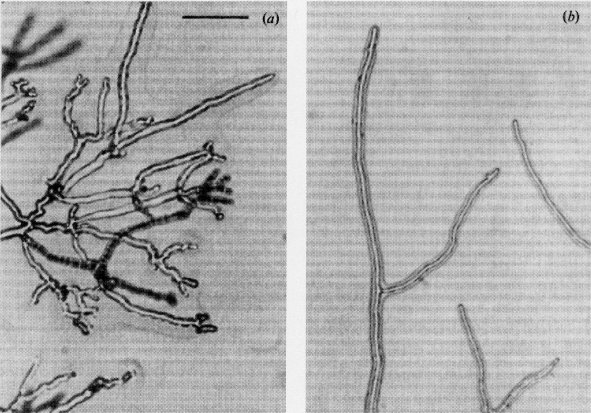
FIGURE 30 Typical morphology of the frost mutant on minimal medium (a) and on minimal medium plus 500 mM Ca2+ (b) The calcium supplement effectively restores wild-type morphology. Bar length = 50 mm. From ref. (533 ), with permission from the Society for General Microbiology.
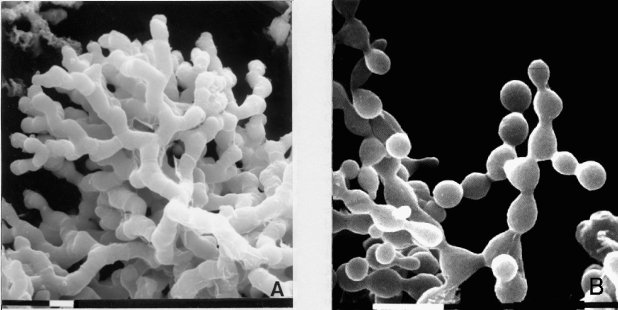
FIGURE 31(A) Aerial hyphae of the mutant fr: frost. (B) Conidiating wild type, for comparison, showing the formation of typical blastoconidia. Bar length = 10 mm. Scanning EM photographs by M. L. Springer. From refs. (1957 ) and (1958 ), with permission from Cold Spring Harbor Laboratory Press.
frq : frequency
VIIR. Between wc-1, for (3%) and oli (<2%), un-10 (1094, 1228, 1309).
Cloned and sequenced: Swissprot PER_NEUCR, PIR S04653, EMBL/Genbank U17073; pSV50 clones 6:5A, 8:3B, 26:2A, 30:1A (55).
A series of frq alleles result in altered periods in the circadian rhythm cycle of conidiation. For reviews, see refs. (144), (148), (558)-(560), (562), (617), (618), (620), (1130), (1224), and (1225) (Fig. 33). frq mutants that are incompletely dominant and that change period length are the following: frq1, 16.5 hr; frq2, 19.0 hr; frq3, 24.0 hr; frq4, 19.0 hr; frq6, 19.0 hr; frq7, 29.0 hr; and frq8, 29.0 hr (561). The recessive allele frq9 shows erratic periodicity; strains carrying this allele have been critical for understanding the gene’s function (1309, 1320, 1321). Scoring is accomplished by zonation in growth tubes or plates using strains that contain bd and preferably csp-1 or csp-2 (618, 1130) (Fig. 32). The presence of csp shortened the period length by about 1hr in one strain tested (529). Temperature compensation and interactions with other loci have been described (617, 711-713). FRQ appears to function in transcription (561), and nuclear localization is essential for function (1236). The levels of frq transcript and FRQ polypeptide show circadian cycles, and this cycling is central to establishing the circadian rhythm (57, 561). Multiple forms of the gene product result from alternative translation initiation sites and time-specific phosphorylation (709, 1214, 1434). The mechanism of temperature resetting has been analyzed (1215).
Until frq was cloned and sequenced, the possibility remained that alleles with different period lengths represented a cluster of different genes. Once allelism was established, the original symbols frq-1, frq-2, etc. were changed to frq1, frq2, etc. To avoid possible confusion, the symbol frq is reserved for this locus (621). However, mutations at numerous loci other than frq have been shown to affect the establishment of the rhythm [e.g., wc-1 and wc-2; (431)] or the period length (e.g., chr and prd). Also, period length and/or temperature compensation are affected by mutations at loci initially recognized on the basis of quite different characteristics, including cel, chol-1, cys-9, fas and phe-1. For a more inclusive list of such loci, see references (558), (561), (1130), and (1133). A mutant called cla proved to be frq7 (397).

FIGURE 32 Diagram of a race-tube culture of the band mutant, expressing a circadian
rhythm of conidiation.
Dark areas are bands of dense conidiation, which occurs at subjective dawn in the cycle. Upper
panel: Side
view of a culture that has completed growth to the end of the tube. Raised areas represent
regions of conidiation
at circadian intervals. Lower panel: Top view of a culture that has grown as far as the present
growth front.
The position of the growth front (vertical lines on the tube) is marked at 24-hr intervals, in red
safelight.
Because the linear growth is nearly constant, the position of the center of the conidial band
relative to the growth-front
marks can be used to determine the time of formation of each conidial band. The period length
then is given by
the number of hours between the centers of successive bands. Adapted from
ref. (618 ),
[Feldman, J. F. (1983). Bioscience 33: 426-431],
with permission, 1983 American Institute of Biological Sciences.

FIGURE 33 The Neurospora frq feedback loop. Circadian time (CT) is a formalism used to compare the properties of circadian rhythms from organisms or strains with different endogenous periods, whereby the period is divided into 24 equal parts with each part defined as 1 circadian hour. By convention, CT0 represents subjective dawn and CT12 represents subjective dusk. From ref. (144 ), where it was adapted from ref. (148 ) [Bell-Pederson, D., N. Garceau, and J. J. Loros (1996). Circadian rhythms in fungi. J. Genet. 75: 387-401.], with permission. Copyright Indian Academy of Sciences, Bangalore.
frq-5 : frequency-5
Changed to prd-1.
fs : female sterile
Infertile as female but fully fertile as the male (fertilizing) parent. No or few functional perithecia are produced. ff (female fertility) and pp (protoperithecia) have been used by other workers to name mutants having a similar phenotype. Female fertility may also be impaired or absent in certain mutants that were named for other traits, e.g., cyt-1, -2, erg, fr, glp-3, gul-3, -4, leu-1, R, ro, sk, so, ssu, ty-1, -2, var-1. Many female-sterile mutants affect vegetative morphology or growth rate. Tests show that different female sterile mutants are blocked at different points in perithecial development (1000). Female sterility appears to have no genetic or functional relationship to mating type (1000). Crosses homozygous for any of the listed fs genes can be made and progeny can be obtained by using a heterokaryon of marked fs and fs+ strains as the female parent (1423, 1424); the same is true for most of the mutants called ff (1000). Numerous additional female-sterile mutants have been isolated that have not been mapped and/or tested for allelism with the mutants listed here (176, 518, 920, 1000).
fs-1 : female sterile-1
I or II. Linked to T(4637) al-1. Unlinked to mat (1423).
Perithecia are absent or infrequent when the mutant is used as female. Fertile as male. Vegetative growth is somewhat stringy, slightly slower, and paler than wild type. Recessive in heterokaryons. Complements fs-2, -3, -4, -5, -6, -n (1423, 1424). Shown in crosses to be nonallelic with fs-2, -3, -5, -n (1423).
fs-2 : female sterile-2
II. Probably linked to fs-1 (1423), cot-5 (14/48) (1582).
No perithecia are formed when the mutant is used as female. Fertile as male. Morphology is abnormal, somewhat colonial. Grows at 25ºC, but not 34ºC. Recessive in heterokaryons. Complements fs-1, -3, -4, -5, -6, -n (1424)
fs-3 : female sterile-3
IL. Left of mat (16%) (1423).
No perithecia are formed when the mutant is used as female. Fertile as male. Vegetative growth is slightly slower and paler than wild type. Recessive in heterokaryons. Complements fs-1, -2, -4, -5, -6, -n (1423, 1424).
fs-4 : female sterile-4
I? Linked to mat (22%) (1423), but may be inseparable from a chromosome rearrangement (1424).
No perithecia are formed when the mutant is used as female. Fertile as male. Complements fs-1, -2, -3, -5, -6, -n. Vegetative growth is slightly slower and paler than wild type. Recessive in heterokaryons (1423, 1424).
fs-5 : female sterile-5
I or II. Probable loose linkage with fs-1 (1423).
Perithecia are absent or rare when fs-5 is used as female. Some strains produce occasional perithecia and ascospores. Fertile as male. Grows slowly, with aerial growth near the agar surface. Cultures turn brown with age (1423). Complements fs-1, -2, -3, -4, -6, -n (1424).
fs-6 : female sterile-6
I or II. Linked to T(4637) al-1. Unlinked to mat (1423).
No perithecia when fs-6 is used as female. Fertile as male. Vegetative growth is slightly slower and paler than wild type. Recessive in heterokaryons. Complements fs-1, -2, -3, -4, -5, -n (1423, 1424).
fs-n : female sterile-n
I. Linked to mat (35%; 45%) and to T(4637) al-1 (1423).
No perithecia when fs-n is used as female. Fertile as male. Vegetative growth is slightly slower and paler than wild type. Recessive in heterokaryons. Three complex segregations in 51 asci suggest two closely linked genes. If so, they do not complement each other, although they complement fs-1, -2, -3, -4, -5, -6 (1423, 1424).
Fsp-1 : Four spore-1
IIR. Right of pe (4%) (1546).
Some of the asci contain four large ascospores rather than the normal eight (Fig. 34). In these asci, ascospores are formed at the four-nucleate stage following meiosis II. The mutant allele is dominant with variable penetrance, depending on genetic background. Ascospores from four-spored asci produce homokaryotic cultures. Some large, heterokaryotic ascospores are found in rare three-spored or two-spored asci. One postmeiotic mitosis is omitted in the four- and three-spored asci, and two divisions are omitted in the two-spored asci. Vegetative morphology is normal (1675). Expression is temperature-sensitive. Asci are four-spored at 25ºC and above (1677). Heterokaryotic large ascospores in three-spored asci have been used in the study of Spore killer to rescue segregants that would not survive as homokaryotic haploids (1674). The developmental basis of the Four-spore mutants of Neurospora crassa is distinct from that of the normally four-spored species Neurospora tetrasperma [for a dominant eight-spored mutant of N. tetrasperma, see ref. (272)].
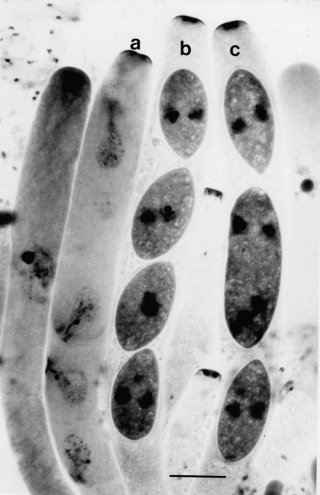
FIGURE 34 Ascus development in a cross parented by the Four spore mutant Fsp-1. (a) Interphase II. The four nuclei with their duplicate spindle-pole-body plaques have failed to undergo mitosis but have lined up on one side of the ascus and initiated ascospore delimitation. (b) An ascus showing four binucleate spores after a single mitosis. No additional mitoses occur after the ascospores become mature. (c) The middle large spore in this three-spore ascus enclosed two of the four meiotic products. Haemotoxylin staining. Bar length = 10 mm. Photographs by N. B. Raju, from ref. (1675 ).
Fsp-2 : Four spore-2
IR. Right of nic-2 (8%) (1677).
In crosses heterozygous for Fsp-2, nearly all asci are four-spored at 16ºC, eight-spored at 25ºC (518). Crosses homozygous for Fsp-2 make four-spored asci at 16 and 20ºC, but a mixture of four- and eight-spored asci at 25ºC . The developmental basis is similar to that of Fsp-1. No asci contain more than four ascospores at any temperature when both Fsp-1 and Fsp-2 are present in a cross, either homozygous or heterozygous (1677). The double mutant has been used to reduce significantly the effort required for tetrad analysis (1602, 1888).
Fsr : 5S rRNA
Almost all of the approximately 100 genes that specify 5S rRNA are widely dispersed in the genome (672, 1893). Sequences flanking the individual 120-bp 5S regions are mostly divergent, but they share an upstream consensus sequence that is required for transcription (1889). Many 5S genes have been partially or completely sequenced, and over 20 have been mapped using RFLPs in their flanking sequences as markers. Most of the 5S genes are of the same sequence type, a, but other isotypes are found, with as many as 15 differences in the 120-bp 5S region (1334, 1339). The variant 5S rRNAs are present in the ribosomes (1892). The symbol Fsr is used both for genes and for pseudogenes.
Fsr-1 : 5S rRNA-1
IR. Linked to al-2 (0T/18 asci; 0/18), un-7, met-6, cr-1 (0/18) (1447). Adjacent to ad-9 (573).
Cloned and partially sequenced (1339).
Specifies a-type 5S rRNA (1339).
Fsr-3 : 5S rRNA-3
IIR. Between preg (5T + 2 double crossovers/18 asci), vma-2 (1T/18 asci) and eas (1T/18 asci), trp-3 (16 + 1 double crossover/41), Fsr-34, Fsr-21 (4/41) (1339, 1447).
Cloned and partially sequenced (1339).
Specifies a-type 5S rRNA (1339).
Fsr-4 : 5S rRNA-4
IVR. Linked to Tel-IVR (1T/18 asci), chs-2, uvs-2 (0/17) (1447). Near Tel-IVR by inclusion in duplications from quasiterminal translocation T(ALS159) and by linkage to T(T54M50) and T(OY337) (1/23) (1339).
Cloned and sequenced: EMBL/GenBank J01247, GenBank NEURG5P.
A 5S rRNA pseudogene (y4); apparently not transcribed (1887).
Fsr-9 : 5S rRNA-9
V. Linked to Cen-V, lys-1 (1T/18 asci), Fsr-16 (0T/14 asci; 0/18) (1447).
Cloned and sequenced (1339).
Specifies b-type 5S rRNA (1339).
Fsr-12 : 5S rRNA-12
IL. Linked to nit-2 (0 or 1/16), vma-4 (0T/18 asci). Left of mat (4T/18 asci) (1447).
Cloned and sequenced: EMBL/GenBank K02478, GenBank NEURGAD.
Specifies d-type 5S rRNA. [Possibly a pseudogene (1339, 1893).]
Fsr-13 : 5S rRNA-13
IVR. Right of trp-4 (0T/18 asci). Linked to vma-8 (0T/18 asci) (1447). Between pyr-1 and cot-1.(1339).
Cloned and sequenced: EMBL/GenBank K02477, GenBank NEURGABP.
Specifies b'-type 5S rRNA (1339, 1893).
Fsr-16 : 5S rRNA-16
V. Linked to Cen-V, lys-1 (1T/14 asci), Fsr-9 (0T/14 asci; 0/18) (1447).
Cloned and sequenced: EMBL/GenBank K02470, GenBank NEURGAAB.
Specifies a-type 5S rRNA (1339, 1893).
Fsr-17 : 5S rRNA-17
IIR. Between eas (6T, 1NPD/18 asci), trp-3 (1/16) and leu-6 (0/16; 9T, 1NPD/18 asci) (1447).
Cloned and sequenced: EMBL/GenBank K02947, K02948, GenBank NEURGAAH, NEURGAAI.
Specifies a'-type 5S rRNA (1339).
Fsr-18 : 5S rRNA-18
IR. Linked to nic-1 (0/18), arg-13 (2T/18 asci) (1447).
Cloned and partially sequenced (1339).
Specifies a-type 5S rRNA (1339).
Fsr-20 : 5S rRNA-20
VR. Right of ro-4 (1/18) (1447).
Cloned and sequenced: EMBL/GenBank K02479, GenBank NEURGAG.
Specifies g-type 5S rRNA (1339, 1893).
Fsr-21 : 5S rRNA-21
IIR. Between arg-12 (1/17) and trp-3 (1/17; 4/41) in the order preg Fsr-3 Fsr-21 trp-3. Linked to Fsr-34 (0/41; 0/18) (1339, 1447).
Cloned and sequenced: EMBL/GenBank NEURGAAE, K02473, also NEURGAAJ, K02949 (OR strains), and NEURGAAK, K02950 (BT strains).
Specifies a-type 5S rRNA (1339, 1893). An allelic difference is present between strains of Beadle-Tatum (BT) and Oak Ridge (OR) ancestry.
Fsr-30 : 5S rRNA-30
IL. Linked to mat (0T/18 asci) (1447).
Cloned and partially sequenced (1339).
Specifies 5S rRNA of uncertain type (1339).
Fsr-32 : 5S rRNA-32
IIL. Between pyr-4 (10/41), Fsr-52 (8/41; 4T/18 asci) and Cen-II (1T/18 asci), arg-5 (2/41) (1339, 1447).
Cloned and sequenced: EMBL/GenBank NEURGAAF, K02474, also NEURGAAL, K02951 (OR strains), and NEURGAAM, K02952 (BT strains).
Specifies a-type 5S rRNA (1339, 1893). An allelic difference is present between strains of Beadle-Tatum (BT) and of Oak Ridge (OR) ancestry.
Fsr-33 : 5S rRNA-33
IR. Linked to lys-4 (0 or 1T/18 asci). Right of his-2 (1 or 2T/18 asci) (1447).
Cloned and sequenced: EMBL/GenBank NEURGATH, M29278 (q), NEURGAZE, M11398 (z-h).
In Oak Ridge and related laboratory wild types, the Fsr-33 locus is occupied by a duplication that includes two heavily methylated pseudogenes (types z and h) that differ by numerous transition mutations (1886, 1890). This region was the first in Neurospora to be recognized as methylated. In numerous wild-collected strains, in wild type Abbot 4A, and in other Neurospora species, the locus contains a single Fsr gene of type q (766, 1891). Sequence comparisons indicate that z and h originated by tandem duplication of an 0.8-kb segment that included q, and that this was followed by ~270 GC-to-AT mutations that are characteristic of RIP (766).
Fsr-34 : 5S rRNA-34
IIR. Between eas (1T/18 asci) and trp-3 (1/18; 4/41). Linked to bli-4 (0T/18 asci), Fsr-21 (0/59) (1339, 1447).
Cloned and partially sequenced (1339).
Specifies a-type 5S rRNA (1339).
Fsr-38 : 5S rRNA-38
Unmapped.
Cloned and sequenced: EMBL/GenBank K02475, GenBank NEURGAAG.
Specifies a-type 5S rRNA (1893).
Fsr-45 : 5S rRNA-45
IIIR. Between ro-2 (3/18) and cat-3 (2T, 1NPD/18 asci), Tel-IIIR (3T, 1NPD/18 asci) (1447).
Cloned and sequenced: EMBL/GenBank K02471, GenBank NEURGAAC.
Specifies a-type 5S rRNA (1339, 1893).
Fsr-50 : 5S rRNA-50
VI. Linked to Bml (0/18), Cen-VI, pan-2 (0T/18 asci) (1447). Between ylo-1 and trp-2 (1339).
Cloned and sequenced (1339).
Specifies a-type 5S rRNA (1339).
Fsr-51 : 5S rRNA-51
IVR. Between arg-14 (2/18), trp-4 (2T/18 asci) and cot-1 (1/18). Linked to bli-3 (0T/15 asci) (1447).
Cloned and sequenced: EMBL/GenBank K02476, GenBank NEURGAB.
Specifies b-type 5S rRNA (1339, 1893).
Fsr-52 : 5S rRNA-52
IIL. Between pyr-4 (2/41), Fsr-32 (3/18; 8/41; 4T/18 asci), Fsr-54 (8/41) and Cen-II (5T/18 asci), arg-5 (10/41) (1339, 1447).
Cloned and sequenced: EMBL/GenBank K02469, GenBank NEURGAAA.
Specifies a-type 5S rRNA (1339, 1893).
Fsr-54 : 5S rRNA-54
IIL. Between pyr-4 (2/41), Fsr-52 (8/41), Fsr-32 (3/18; 0/41) and arg-5 (10/41) (1339, 1447).
Cloned and partially sequenced: EMBL/GenBank K02472, GenBank NEURGAAD.
Specifies a-type 5S rRNA (1339, 1893).
Fsr-55 : 5S rRNA-55
IIR. Between Cen-II, arg-5 (1T/18 asci; 2/39), con-6 (1/18) and preg (4/39; 1T + 1 double crossover/18 asci). Linked to cya-4 (0T/18 asci) (1339, 1447).
Cloned and partially sequenced (1339).
Specifies a-type 5S rRNA (1339).
Fsr-62 : 5S rRNA-62
IVR. Linked to Fsr-63 (0/18), mtr (0/18), arg-14 (1/18) (1447).
Cloned and partially sequenced (1339).
Specifies b-type 5S rRNA (1339).
Fsr-63 : 5S rRNA-63
IVR. Between Cen-IV (2T/15 asci) and pyr-1 (1T/18 asci), mtr (0/18), arg-14 (1/18). Linked to Fsr-62 (0/18) (1447).
Cloned and sequenced (1339).
In Abbott 12a and Abbott 4A strains, the locus is occupied by an a-type 5S rRNA gene. In Oak Ridge strains, however, the 5S rRNA gene is interrupted by a 1.9-kb transposon, Punt. The copy of Punt in Fsr-63 and the surrounding 5S rDNA sequences are methylated and have the earmarks of RIP. This pseudogene is called y63 (1266).
fz : fuzzy
Unmapped.
Morphology is abnormal. Identified as one component of the multiple mutant combination that results in the cell-wall-less “slime” phenotype (607).
Return to the 2000 Neurospora compendium main page