G : Gulliver
Changed to gul: gulliver.
Gamma-tubulin
See tbg.
gap : gap
IL. Between mat (6%) and Cen-I (4%) (1200).
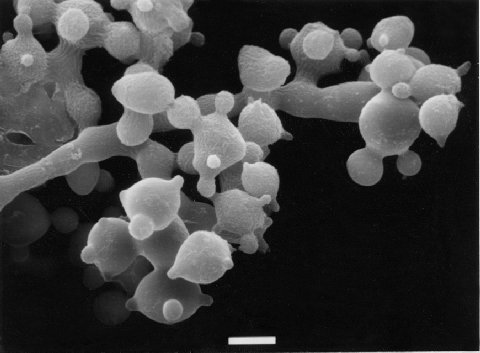
Conidia are formed in a few scattered clusters on long hyphae otherwise devoid of conidia. Photograph (1200). (Stock lost.)
gch-1 : GTP cyclohydrolase-1
Unmapped.
Cloned and sequenced: Swissprot GCH1_NEUCR, EMBL/GenBank Z49758, Genbank NCGTPCHI.
Encodes GTP cyclohydrolase I (EC 3.5.4.16) (GTP-CH-I) (1253).
gdh-1 : glutamate dehydrogenase-1
VIL. Linked to tom22 (0T/16 asci), nuo19.3 (9T/18 asci) (1447). Apparently not linked in III as originally reported (1027).
Cloned and sequenced: EMBL/Genbank L20497, PIR S66039, EMBL NCNADSPEC, GenBank NEUNADSPEC
Structural gene for NAD-specific glutamate dehydrogenase (EC 1.4.1.2) (1027) (Fig. 49).
gla-1 : glucoamylase-1
Unmapped.
Cloned and sequenced: Swissprot AMYG_NEUCR, EMBL/GenBank X67291, GenBank NCGLA1, PIR S36364, S13710, S13711; EST NM6B8.
Structural gene for the major extracellular glucoamylase (EC 3.2.1.3) by homology and by RIP knock-out. Mutants have reduced starch halos and reduced growth on starch as the sole carbon source (2005). Residual glucoamylase in the null mutant may be due to gla-2.
gla-2 : glucoamylase-2
IL. Linked to mat (3/68), sor-4 (£10 kb), his-3 (3/65) (576).
Cloned and partially sequenced: Orbach-Sachs clone X10E5 (576).
Structural gene for extracellular glucoamylase (EC 3.2.1.3) by homology. Little sequence homology to gla-1. Homologous to a glucoamylase from Schwanniomyces occidentalis (576).
glm : glutamine
Symbol changed to gln.
gln-1 : glutamine-1
VR. Linked to inl (2%, probably to the right) (1707).
Requires glutamine (1707). Probably a glutamine synthetase structural gene (447, 1376, 1781) (Figs. 49 and 50). Sensitive to chlorate on both ammonium and glutamate; resistant to chlorate on glutamine (565). Inactivation by oxidation (8); alterations in activity during development are associated with hyperoxidation (2097). NADPH-nitrate reductase, NAD(P)H-nitrite reductase, and uricase are freed from repression by ammonium or glutamate but not by glutamine in the gln-la mutant (564, 567, 1648, 2184). A gln-1b allele, defective in the b-subunit, is more derepressed than mutant R1015 (gln-1a), which is defective in the α-subunit (565, 1650). Glutamine synthetase can be found in an octameric form composed of either β-subunits exclusively or b-subunits plus a-subunits and in a tetrameric form consisting exclusively of a-subunits (1376). The a- and b-subunits are encoded by different mRNAs (1140). The enzyme is allosterically inhibited by alanine, glycine or serine, and a b-subunit mutant sensitive to glycine has been isolated (873). For interaction with am, see ref. (939). Dominant in heterokaryons (446). Regulated by nit-2 and nmr-1 (269). The gln-1bR8 strain has been used to obtain a mutant with altered L-amino acid oxidase regulation (271). Called glm (1707).
gln-2 : glutamine-2
Unmapped. Unlinked to gln-1.
Affects glutamine synthetase a-polypeptide (270). Transferase activity drastically is reduced and that of glutamine synthetase is slightly reduced. Resistant to a-methyl-DL-methionine-SR-sulfoximine, an inhibitor of glutamine synthetase activity (270).
glp : glycerol phosphate
Mutants with altered ability to use glycerol as a carbon source. For pathways of glycerol utilization in various organisms, see the diagram in ref. (2160) or (2098). Scored in slants on minimal synthetic cross medium (2208) with 2% glycerol vs. 2% sucrose as the carbon source (2160). The poor growth of wild type on glycerol medium is markedly improved by adding 0.5% L-asparagine and 100 mg/ml ascorbic acid (361). Symbol changed from gly.
glp-1 : glycerol phosphate-1
IR. Probably between ad-9 (2%) and nit-1 (11%) (890, 1478).
Unable to use glycerol as the sole carbon source (1478). Can use dihydroxyacetone or glyceraldehyde (525). Probably regulatory. Deficient in inducible glycerol kinase under normal conditions (890, 1481); wild-type levels of normal enzyme are induced by cold or by deoxyribose with some, but not with all, alleles (525, 890). Glycerol transport is normal (525). Fine-structure map (526). Scored on slants of minimal synthetic cross medium (2208) with 2% glycerol vs. 2% sucrose as the carbon source (2160). Poor growth of wild type on glycerol medium is markedly improved by adding 0.5% L-asparagine and 100 mg/ml ascorbic acid (361). Called gly, gly-u.
glp-2 : glycerol phosphate-2
IIR. Between T(ALS176), arg-5 (8%) and T(NM177)L, pe (7%). Linked to aro-3 (3%) (527, 1578).
Unable to use glycerol, dihydroxyacetone, or glyceraldehyde as the sole carbon source (525, 527). Lacks both mitochondrial and cytosolic flavin-linked glycerol-3-phosphate dehydrogenase (EC 1.1.99.5) (527). Scored on slants of minimal synthetic cross medium (2208) with 2% glycerol vs. 2% sucrose as the carbon source (2160). Poor growth of wild type on glycerol medium is markedly improved by adding 0.5% L-asparagine and 100 mg/ml ascorbic acid (361). Three independent isolates are ropy-like in vegetative morphology but are female-fertile, unlike most ropy mutants (419, 527). A report of complementation groups at this locus (420) is an error (417). Fine-structure map (526). Called gly-2.
glp-3 : glycerol phosphate-3
Allelic with ff-1.
glp-4 : glycerol phosphate-4
VI. Between ylo-1 (1%; 6%), ad-1 (0; 2%) and rib-1 (3%; 4%), pan-2 (4%; 6%) (2160).
Unable to use glycerol as the sole carbon source (2160). Uses dihydroxyacetone or glyceraldehyde (525). Lacks both inducible and constitutive glycerol kinase (EC 2.7.1.30) (2160), but there is some doubt that these are two different enzymes [ref. (417), based on ref. (526)]. Scored on slants of minimal synthetic cross medium (2208) with 2% glycerol vs. 2% sucrose as the carbon source (2160). The poor growth of wild type on glycerol medium is improved markedly by adding 0.5% L-asparagine and 100 mg/ml ascorbic acid (361). A revertant has altered kinetic properties (526). Allele G660 originated in Neurospora tetrasperma and was introgressed into Neurospora crassa (155, 2160). Fine-structure map (526). See Fig. 18.
glp-5 : glycerol phosphate-5
I. Left of cr-1 (15%) (2160).
Unable to use glycerol as the sole carbon source. Lacks glyceraldehyde kinase (2160), but the significance of this is uncertain because of the findings of ref. (2098). Scored on slants of minimal synthetic cross medium (2208) with 2% glycerol vs. 2% sucrose as the carbon source (2160). Poor growth of wild type on glycerol medium is improved markedly by adding 0.5% L-asparagine and 100 mg/ml ascorbic acid (361). Allele M1051 originated in Neurospora tetrasperma and was introgressed into Neurospora crassa (155, 2160).
glp-6 : glycerol phosphate-6
V. Left of inl (30%) (1654).
Deficient in NAD-linked glycerol-3-phosphate dehydrogenase (EC 1.1.1.8). Called 42-94 (914, 1654).
glt : glycyl-leucyl-tyrosine resistant
Unmapped.
Unable to transport oligopeptides necessary to support the growth of specified amino acid auxotrophs (2236). Has only 10% of wild-type uptake rate (2237). (The oligopeptide uptake system transports tri-, tetra-, and pentapeptides, but not di- or higher than pentapeptides.) However, whereas a leu-2 gltR strain is unable to use oligopeptides as a source of leucine when ammonium nitrate is provided as a nitrogen source, the double mutant but not the leu-2 single mutant can use small peptides as a nitrogen source, apparently because of an induced extracellular peptidohydrolytic activity (2238). Originally obtained using tys by selecting mutants resistant to glycyl-L-leucyl-L-tyrosine but still sensitive to tyrosine (2236). See ref. (2232) for a review of peptide uptake.
gluc-1 : β-glucosidase-1
IIIR. Linked to dow (10%) (579).
Activity of the thermostable aryl-b-glucosidase is reduced to 10% of wild type (582) in one allele and to <1% in a second-step mutant then called gluc-2, which showed 0/200 recombination with the original mutant and is probably allelic (581). Low activity is dominant in heterokaryons (1246). Selected by p-nitrophenylglucoside-staining reaction (580). Scored either by breakdown of the b-glucoside esculin (0.01%) measured by fluorescence at pH 5.5 (582) or by precipitation of ferric ammonium citrate (0.1%) by esculetin (2 days, 25ºC) (579).
gluc-2 : β-glucosidase-2
Probably allelic with gluc-1.
gly : glycerol utilization
Symbol changed to glp.
gna-1 : guanine nucleotide-a-1
IIIR. Linked to ro-2 (0/17) (2130).
Cloned and sequenced: Swissprot GBA1_NEUCR, EMBL/GenBank L11452, U56090, EMBL NCGPROTAA, Genbank NEUGPROTAA; Orbach-Sachs clone X9F04, G9C03.
Encodes guanine nucleotide-binding protein a-1-subunit (2130). The predicted amino acid sequence shows significant homology to the Gi family found in higher organisms. The null mutant shows female infertility, osmotic sensitivity, reduced hyphal growth rate, and defective macroconidiation (972), has decreased intracellular cAMP levels, and is more resistant to some environmental stresses than wild type (973, 2260). Mutants that are predicted to maintain an activated state of GNA-1 have increased intracellular cAMP levels, overproduce aerial hyphae, and are more sensitive to environmental stresses than wild type (2260). Null-mutant extracts have reduced Mg2+-dependent adenylyl cyclase and cAMP phosphodiesterase activity. Mg2+-dependent adenylyl cyclase activity in wild-type extracts can be inhibited by using antibody against GNA-1 (973).
gna-2 : guanine nucleotide-a-2
VR. Linked to inl (0/18) (2130).
Cloned and sequenced: Swissprot GBA2_NEUCR, EMBL/GenBank L11453, EMBL AF004846, Genbank NEUGPROTAB; EST NP3A7; Orbach-Sachs clones X23E01, X24E12.
Encodes guanine nucleotide-binding protein a-2-subunit (2130). Expressed in both vegetative and sexual stages. The null mutant has no obvious phenotype. Mutants containing predicted GTPase-deficient (activated) alleles of gna-2 have increased aerial hyphae and reduced conidial germination, but normal female fertility. The null mutant has normal levels of Mg2+-dependent adenylyl cyclase activity but reduced levels of cAMP phosphodiesterase activity (973). gna-1, gna-2 double mutants are more impaired than gna-1 strains in osmotic sensitivity and female fertility (80).
gna-3 : guanine nucleotide-a-3
Probably IV, near uvs-2 (199)
Cloned and partially sequenced.
Encodes guanine nucleotide-binding protein a-3-subunit (199).
gnb-1 guanine nucleotide-b-1
IIIR. Linked to con-7, trp-1 (0/18) (199)
Cloned and partially sequenced.
Encodes guanine nucleotide-binding protein b-subunit . The null mutant is female-sterile and has reduced levels of GNA-1 protein under some growth conditions (199).
IIIR. Linked to con-7, trp-1 (0/18) ( gpd-1 : glyceraldehyde-3-phosphate dehydrogenase-1
IIR. Linked to arg-12 (0/18) (1777), vma-2 (1T/18 asci). Between nuo78, preg, cit-1 (5T/18 asci) and eas (3T/18 asci) (1447).
Cloned and sequenced: Swissprot G3P_NEUCR, EMBL/Genbank U56397, U67457, U56379; EST NC2H9.
Structural gene for glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.12) (1777, 1910). RNA peaks late in the circadian night and enzyme activity peaks in late morning; gene regulation other than circadian control has not been found (575, 1910). Used in a study of phylogenetic relationships among homothallic and heterothallic species of Neurospora and Sordaria (1639). Also called ccg-7.
gpi-1 : glucose phosphate isomerase-1
IVR. Linked to ad-6 (10%) (1401).
Lacks glucose phosphate isomerase (phosphohexoisomerase) (EC 5.3.1.9). Grows slowly and colonially on glucose or sucrose. Unable to use fructose, but growth on glucose is stimulated by added fructose. Growth is enhanced in double mutants with either sor-4(T9) or pp. Allele T66M37 was originally called gpi-2 (1401).
gpi-2 : glucose phosphate isomerase-2
Used for gpi-1 allele T66M37.
gran : granular
VR. Between pab-2 (1%; 8%) and his-6 (8%; 27%) (1582, 1592). Linked to pl (0/75).
Delicate granular conidiation, with conidia adherent rather than powdery (1585). Exaggerated major and minor constrictions (1958) (Fig. 35). Sparsely branched hyphae (719). Morphologically distinct from pl. Cell-wall peptides are reduced in amount (2249).

FIGURE 35 The mutant gran: granular, showing late budding. Bar length = 10 mm. Scanning EM photograph from M. L. Springer.
grey : grey
IVR. Linked to cot-1 (4%) (932).
Produces grey conidia (microconidia?) in the presence of cr-1 (932).
grg-1 : glucose-repressible gene-1
Allelic with ccg-1.
grn : guanyl-specific RNase
Unmapped.
Sequenced: Swissprot RNN1_NEUCR, PIR NCNCT1.
Specifies guanyl-specific ribonuclease N1 (EC 3.1.27.3) (2038).
grp78 : glucose-regulated protein 78
I. Linked to mat, Fsr-30 (0T/18 asci) (1447).
Cloned and sequenced: EMBL/GenBank Y09011, GenBank HCHSP70GR.
Encodes a glucose-regulated and heat-shock-inducible protein of the hsp70 family, homologous with Saccharomyces KAR2/GRP78. The mRNA level is low in dormant conidia and then increases at germination, low in young aerial hyphae and then increasing (811). Called hsps-2.
gs : gamma sensitive
The symbol gs was used for a series of mutants sensitive to g-radiation (1316). These were not mapped or preserved. They are listed in ref. (1596) but are not included here.
gs-1 : glucan synthase-1
Allelic with cot-2.
gsp : giant spore
IL. Left of mat (10%) (1966).
Some asci contain a single giant ascospore and others have two very large ascospores or four double-size ascospores, whereas some asci contain eight ascospores. Proportions of these types vary on different crossing media. The giant ascospores have multiple germination pores. The mutant ascus phenotype is recessive. Vegetative morphology is normal, but growth is weak (1148, 1966).
gsy-1 : glycogen synthase-1
Unmapped.
Cloned and sequenced: EMBL/GenBank AF056080.
Encodes glycogen synthase (EC 2.4.1.11, UDP-glucose-glycogen glucosyltransferase) (2075).
gtp-1 : guanine triphosphate binding-1
Unmapped.
Cloned: Orbach-Sachs clone G11G09 (1267).
The predicted protein shows similarity to the product of S. pombe gtp1 and vertebrate DRG (“developmentally regulated gene”) protein. This class of proteins is found in organisms from bacteria (obg in Bacillus subtilis, an essential gene possibly involved in regulating DNA replication) to fungi (gtp1 in S. pombe, with no obvious mutant phenotype) to animals (DRG, induced in developing brain tissue). Contains a GTP-binding domain shown to be critical to function.
gua : guanine
The guanosine-specific branch of the purine biosynthetic pathway diverges with the conversion of inosine 5¢-monophosphate to xanthosine 5¢-monophosphate. In yeast there are at least four genes encoding four distinct isozymes for this function, IMP dehydrogenase (EC 1.1.1.205). In Neurospora, gua-2 definitely is blocked in this enzyme, and gua-1 may specify an inducible isozyme. No mutant is known yet for the subsequent step, conversion of xanthosine-5¢-P to guanosine-5¢-P by guanosine-5¢-monophosphate synthase (EC 6.3.5.2). See Figs. 5 and 62.
gua-1 : guanine-1
I. Linked to arg-3 (8%) (2278), probably between his-2 (3%) and cr-1 (3%) (1582).
Requires guanine. Deficient in IMP dehydrogenase (EC 1.1.1.205) (10% of wild type in allele OY301) (767) (Fig. 5). Inhibited (competitively) by adenine and by complex complete medium. Adapts phenotypically after several days and grows up on minimal or complete medium, but retains the requirement on subculture. Adenine prevents or decreases adaptation. Guanosine is preferred to guanine as a supplement because of greater solubility. Best scored at 2 and 3 days on slants of minimal + 1 mg/ml adenine vs minimal + 0.2 mg/ml guanosine (2278).
gua-2 : guanine-2
IVR. Unlinked to gua-1 (25% prototrophs in intercross (768). Linked to cot-1 (5%) (767).
Requires guanine. No IMP dehydrogenase activity (EC 1.1.1.205) (Fig. 5). Slow growth, poor conidiation. Does not adapt to grow on minimal medium (767, 768)
Guest (Transposable element)
Cloned and sequenced: Orbach/Sachs cosmid G36F (2274).
A DNA transposable element discovered as a 98-bp fragment in wild type ST74A, in the region between his-3 and cog. The element is flanked by a 3-bp duplication of target sequence and has terminal inverted repeats (TIRs) similar to those of non-retrotransposon-type elements in other organisms. Multiple copies of sequences that are similar, but not identical, to the TIR are present elsewhere in the genome (2274, 2275).
gul : gulliver
This name was given to suppressors of cot-1 that give large colonies at restrictive temperatures, where unsuppressed cot-1 forms tiny colonies (1714). Scorable in the presence of cot-1 at 34oC, 2 days after the transfer of small inocula to solid medium. Of 36 independent gul mutants, 25 were gul-1 alleles (2079).
gul-1 : gulliver-1
VR. Between am-1 (<0.01%; <1%) and ace-5 (<1%) (1121, 1941).
Cloned (217).
Modifies the colony size of cot-1 at restrictive temperatures (1714, 2079). cot-1; gul-1 colonies exceed 20 mm in diameter after 60 hr at 33ºC, compared to 1 mm for cot-1; gul-l+ (2079). Female fertile, with viable ascospores. Dominant in heterokaryons (217). Recombination within gul-1 is unaffected by rec-3, which acts on the nearby am-1 locus (1941). Called G (1714).
gul-2 : gulliver-2
Unmapped.
Modifier of the colony size of cot-1 at restrictive temperatures. Phenotype similar to gul-1 (2079).
gul-3 : gulliver-3
IVR. Linked to cot-1 (10%), pyr-2 (7%) (2079).
Modifier of the colony size of cot-1 at restrictive temperatures. Female sterile. gul-3 ascospores are black but inviable. gul- progeny can be obtained, however, from gul+/gul- pseudo-wild disomic ascospores. Reported unable to make heterokaryons (2079).
gul-4 : gulliver-4
VII. Linked to nic-3 (17%) (2079).
Modifier of the colony size of cot-1 at restrictive temperatures. Resembles gul-3 (2079).
gul-5 : gulliver-5
VI. Linked to trp-2 (10%) (2079).
Modifies the colony size of cot-1 at restrictive temperatures. Female-fertile. gul-5 ascospores are black, but inviable (2079).
gul-6 : gulliver-6
Unmapped. Unlinked to cot-1 (IVR), inl (VR), nic-3 (VIIL), gul-5 (VI), or gul-2 (2079).
Modifier of the colony size of cot-1 at restrictive temperatures. Said to resemble gul-3 (2079). However, ascospore ripening and recovery from ascospores have been found to be good (1582).
Return to the 2000 Neurospora compendium main page