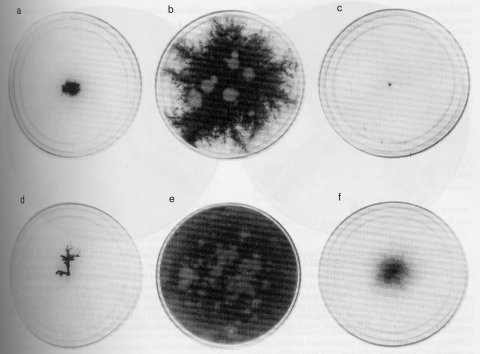
hah : high aerial hyphae
VR. Linked to inl (13%) (1404).
Obtained as a suppressor of the colonial growth and low cAMP level of cr-1. High aerial hyphae are formed on agar medium. The double mutant cr-1; hah conidiates, but the single hah mutant does not (1399, 1404). See also ref. (1020).
hak-1 : high-affinity potassium transporter
Unmapped.
Cloned and sequenced: EMBL/GenBank AJ009759, GenBank NCR9759.
Encodes a K+-H+ symporter homologous to the HAK potassium transporter of Schwanniomyces occidentalis. The potassium transporter is inducible when cultures are K+-starved (842). See also trk-1.
ham : hyphal anastomosis
Allelic with so (809a).
has : hydroxamic acid sensitive
Unmapped. Unlinked to azs (595).
Lacks the salicylhydroxamic acid (SHAM)-sensitive respiratory pathway. Cannot produce the hydroxamate-sensitive respiratory pathway when grown in the presence of chloramphenicol. Grows slowly in the presence of antimycin A (595). The double mutant has; azs is unable to grow in the presence of antimycin A, whereas wild type and has+; azs grow well. The has; azs strain (called ANT-1: antimycin-sensitive) was used to obtain mutants resistant to oligomycin (596) and mutants deficient in succinate dehydrogenase (590).
hbs : homebase
II. Linked to Fsr54 (0 or 1/18). Left of con-6 (1 or 2/18), arg-12 (3 or 4/18) (1447).
An anonymous DNA fragment discovered when it was inserted as a duplicate copy in the am locus (strain 77s15) (1055, 1060).
hda-1 : histone deacetylase-1
Unmapped.
Cloned and sequenced (545).
The sequence resembles HDA1 in Saccharomyces (545).
hda-2 : histone deacetylase-2
IR. Linked to arg-13 (2T/18 asci) (545).
Cloned and sequenced (545).
The sequence resembles HOS2 in Saccharomyces and phd1 in S. pombe (545).
hda-3 : histone deacetylase-3
Unmapped.
Cloned and sequenced (545).
The sequence resembles RPD3 in Saccharomyces (545).
helper-1
Used for the strain am1ad-3B cyh-1 (FGSC No. 4564), which is able to form vigorous wild-type heterokaryons with OR-compatible mutant strains of either mating type (1564). Because mat a allele am1 is inactive, the helper-1 component of such a heterokaryon is a passive partner when the heterokaryon is used as parent in a cross. Only nuclei of the active partner participate in karyogamy, meiosis, and the production of sexual progeny. The helper-1 strain has proved useful for crossing otherwise infertile or poorly fertile strains (1565), for rescuing and sheltering inviable or unstable genotypes (114, 1576), and for determining whether strains of undetermined mating type are OR-heterokaryon-compatible.
Used for the strain am1ad-3B cyh-1 (FGSC No. 4564), which is able to form vigorous wild-type heterokaryons with OR-compatible mutant strains of either mating type
het : heterokaryon (vegetative) incompatibilityIf two strains carry different alleles at one or more het loci, they are unable to form stable heterokaryons (715, 716). Incompatibility due to het genes is strictly vegetative; fertility of crosses is not reduced when parent strains are vegetatively incompatible. Vegetative incompatibility may be manifested in several ways: (a) Failure to form stable heterokaryons (715, 889), best seen using complementing auxotrophic or other forcing markers. (b) Cell death following the fusion of unlike hyphae (721) or after the microinjection of cytoplasm or extracts into unlike strains (2226). The hyphal segments involved are sealed off and die; incompatible nuclei do not migrate through septal pores into adjoining cells. Microinjection implicates proteins in the killing reaction (2213, 2226). (c) Abnormal growth, morphology, pigmentation, and cell death in colonies of meiotically generated partial diploids that are heterozygous for one or more het genes (1421, 1475, 1560) (Fig. 36). Duplication-generating chromosome rearrangements enable individual het genes to be identified, mapped, and characterized one at a time in an otherwise haploid background using the abnormality of heterozygous duplication progeny as a criterion (1560, 1578). Cloning of het genes has also made use of the abnormality of heterozygous transformants (1346, 1934). (d) Occurrence of a barrage reaction following the confrontation of vegetatively incompatible strains (776, 780, 1597). When the interacting strains are of opposite mating type, het incompatibility is manifested by the appearance of two rows of perithecia separated by a clear zone within which killing occurs (Fig. 37). The barrage is seen most clearly when strains are used that do not form macroconidia.
het-c, -d, -e, and -i were detected in laboratory strains, using heterokaryon tests (715, 1630, 2225). Discovery of het-5 through het-10 was based on the abnormality of duplication progeny when a series of duplication-generating rearrangements were crossed with strains from nature (1421). het genes are polymorphic in natural populations of Neurospora crassa (1425). The mating-type idiomorphs A and a also act as het genes in N. crassa (134, 721, 1475, 1629). The mat A/mat a vegetative incompatibility reaction depends on the presence of a functional tol+ allele (977, 1466). Genetic differences at other loci can affect the vigor, stability, and speed of formation of heterokaryons (51, 488, 980). Dominant suppressors of the incompatibility reaction have been reported that affect one or more het loci (51). Rockefeller-Lindegren (RL) wild types are het-C, het-D, het-E (2225), and het-I (2222). St. Lawrence 74A and Oak Ridge (OR) wild types are het-C, het-d, het-e (2225), and het-i (1630, 2222). By definition, OR strains are hetOR for het loci other than these four (1421), e.g., het-5OR. Tester strains are available for identifying and scoring the known het genes (700, 1588). The Wilson-Garnjobst testers for het-c, -d, and -e are complicated by differences at another locus or loci affecting heterokaryon formation (980, 2223). For map locations of het genes and the rearrangements used for testing, see Fig. 1 in ref. (1588). The inactive-mating-type helper strain am1ad-3B cyh-1 (779) is useful for determining whether a strain is het-compatible with OR strains (1564). het gene differences affect the transmission of mitochondrial plasmids between strains, both vegetatively (509) and in sexual interactions (508). Vegetative incompatibility is reviewed in refs. (742), (1176), and (1568).

FIGURE 36 Abnormal growth of duplication (partial diploid) strains that are heterozygous at different het loci. Differences in morphology and severity of inhibition are apparent. (a) mat A/mat a from a cross heterozygous for In(H4250). (b) het-C/het-c from T(NM149) het-C X het-c. (c) het-E/het-e from T(T54M50) het-e X het-E. (d) het-8OR/het-8AD from T(T39M777) het-8OR X het-8AD. (e) het-9OR/het-9PA from T(AR209) het-9OR X het-9PA. (f) het-10OR/het-10AD from T(5936) het-10OR X het-10AD. Strains were inoculated onto minimal agar medium supplemented with phenylalanine + tyrosine, without which brown pigment does not form. Pale-colored areas on the brown-pigmented surface in (b) and (e) are patches of noninhibited growth resulting from breakdown of the duplication. From ref. (1421 ), with permission from the Genetics Society of America. For additional photographs of het-incompatible heterozygous partial diploids, see refs. (1475 ) (mat) and ( 979 ) (het-c, het-6).
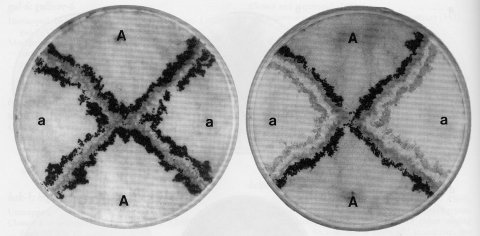
FIGURE 37 Barrage formation as a manifestation of vegetative (heterokaryon) incompatibility. Strains of opposite mating type, mat A and mat a, were inoculated to crossing medium in alternate quadrants of the petri dish at the positions marked. When unlike strains come together, a clear zone of inhibition is formed and lines of perithecia develop on each side of this barrage. Each strain acts as the maternal parent of perithecia on its own side of the barrage, and the strain on the opposite side acts as the fertilizing parent. This is shown in the plate to the right, where the mat a parent carried per allele PBJ1. The perithecial color mutant blocks formation of black pigment in the maternally generated perithecial wall. The nonblack perithecia, which become orange as carotenoids develop, were fully as numerous as the black perithecia opposite them (1569 ). Parents in these tests all carried a mutant fluffy allele, which improves visibility by eliminating macroconidia. Photographs from D. D. Perkins.
het-c : heterokaryon incompatibility-c
IIL. Between cot-5 (3%) and Pad-1, pyr-4 (1%), ro-3. In a common cosmid with Pad-1. Included in duplications from T(NM149) and T(AR179), but not from T(P2869) (924, 1109).
Cloned and sequenced: EMBL/GenBank L77234, EMBL NCHETC, GenBank NEUHETC; Orbach-Sachs clone G22H5.
Specifies a polypeptide that contains a hydrophobic sequence, a leucine-rich domain, and a glycine-rich domain. Allele specificity is due to a highly variable domain of about 40 amino acid residues (1797). Stable heterokaryons are not formed by strains with het-c alleles that differ in specificity (715, 716). Heterozygous duplications show inhibited “brown flat” morphology, spreading to cover a slant but not conidiating (1552, 1560) (Figs. 36 and 37). Two alleles found in laboratory strains were originally called het-C and het-c (715). Following the discovery and demonstration of a third allele, het-cPA, the symbol het-C was changed to het-cOR and the symbol het-c was changed to het-cEM (1797). Three specificity types have been recognized and shown to be polymorphic in nature. These are designated het-cOR, het-cGR, and het-cPA. (het-cEM is functionally identical to het-cGR.) Polymorphisms in other Neurospora species suggest that the known het-c alleles originated prior to divergence of the species (1797). Differences at het-c are more effective than those at het-d, het-e, or mat in preventing the transmission of mitochondrial plasmids between strains with different allele specificity (509). Called c.
het-d : heterokaryon incompatibility-d
IIR. Right of fl (25%) (715). Included in duplications from T(ALS176) (1562) and T(OY337) (1578).
Stable heterokaryons are not formed by strains het-D + het-d (715, 716); het-D/het-d duplications show inhibited spreading growth on slants, with fine subsurface hyphae and no conidia. These are distinguishable from het-C/het-c duplications, which have coarser texture (1562).
het-e : heterokaryon incompatibility-e
VIIL. Between cya-8, T(ALS179) and nic-3 (28%) (1578, 2225). Included in duplications from T(T54M50).
The killing reaction following the fusion of het-E and het-e mycelia is more rapid and severe, and growth inhibition in het-E/het-e duplication strains is more severe than for the vegetative incompatibility mediated by different alleles at het-c or mat (1560, 2225) (Fig. 36).
het-i : heterokaryon incompatibility-i
I or II. Linked to T(IR;IIR)4637 al-1 (1630). Second-division segregation in five of eight asci (2223).
Recognized by cessation of growth of forced heterokaryons. In heterokaryons between the strains used by Pittenger (1630), het-i nuclei were eliminated and het-I nuclei were retained if the initial frequency of het-I exceeded 30%. When more than 70% of nuclei were het-i, growth continued without a change of ratio. In heterokaryons between the RL strains used by Calligan and Wilson, nuclei called Hi are eliminated from HI + hi heterokaryons regardless of the starting ratio (2223). The genes Hi and hi (“heterokaryon instability”) from RL strains are believed to be alleles of het-I and het-i, which initially were called I and i.
het-5 : heterokaryon incompatibility-5
IR. Between R, T(MD2)L and T(MD2)R, T(NM169d), un-18 (978, 1578).
Vegetative incompatibility, initially recognized by inhibited duplications, was confirmed using heterokaryon tests (1421, 1425). Can be scored in duplication progeny from crosses that are heterozygous for either T(MD2) or T(NM103) (1588).
het-6 : heterokaryon incompatibility-6
IIL. Between T(AR18)L, hsp70, un-24 (0/222; 14 kb), and T(AR18)R; hence between cys-3 and T(P1869), cot-5 (1421, 1935).
Cloned and sequenced: GenBank AF206700 (OR), AF208542 (PA); Orbach-Sachs clones G8G1, X14C1.
Duplications heterozygous for het-6 are severely inhibited. Escape from inhibition occurs by deletion of a segment carrying one of the het alleles from the partial diploid produced by T(AR18) (1938). Vegetative incompatibility was recognized by the production of inhibited duplication progeny when strains from nature were crossed by T(AR18), T(P2869), or T(NM149) (1421, 1578). Confirmed by heterokaryon tests (978). Alleles at het-6 and un-24 are in linkage disequilibrium, with no coupling-phase recombinant in strains from natural populations (1351).
het-7 : heterokaryon incompatibility-7
IIIR. Between T(D305)L, T(UK8-18)R and T(AR17)L. Hence, right of un-17, nit-7 and left of dow (1578). Linked to cyt-19 (5%; 9%) (1377). Included in duplications from T(D305) (1421).
Vegetative incompatibility was recognized by the production of inhibited duplication progeny in crosses of wild strains by T(D305). No heterokaryon tests.
het-8 : heterokaryon incompatibility-8
VIL. Between ser-6 (8%) and ad-8 (3%; 12%), T(T39M777) (1172, 1421, 1425).
Vegetative incompatibility initially was recognized by the production of inhibited duplications from crosses of wild strains by T(T39M777) (Fig. 36) and was confirmed using heterokaryon tests (1425). Crosses of T(T39M777) with isolates from nature indicate that there are either three alleles (het-8OR, het-8HO, and het-8PA) or another polymorphic het locus closely linked to het-8 (924).
het-9 : heterokaryon incompatibility-9
VIR. Between T(AR209) and T(OY329)L; hence, between Cen-VI and trp-2 (1578).
Vegetative incompatibility was recognized by the production of inhibited duplications from crosses of wild strains by T(AR209). No heterokaryon test. Photograph of heterozygous duplication colony (1421).
het-10 : heterokaryon incompatibility-10
VIIR. Between dr, T(5936) and Tip-VIIR. Included in duplications from T5936) (1421, 1578).
Recognized by the production of inhibited duplications in crosses of wild strains by T(5936) (1421). No heterokaryon test (Fig. 36).
het-11 : heterokaryon incompatibility-11
Unmapped.
Identified and cloned using duplications of random sequences obtained by ectopic integration of cosmids from a library (1345, 1346).
het-12 : heterokaryon incompatibility-12
V (1492).
Identified by the slow growth of transformants obtained using a linkage group V cosmid. Thought at first to be a suppressor of het-c-mediated vegetative incompatibility and then reinterpreted as containing an independent functional het gene (1492, 1493). Published information is insufficient to establish the validity of the postulated het-12 locus.
hgu-4 : histidylglycine uptake-4
VR. Between cyh-2 (7%) and ure-2 (10%) (2230).
Unable to use L-histidylglycine to support his-6 (2235). Reduced by approximately 50% in its ability to transport most of the amino acids tested. Resistant to many amino acid analogs (2230).
hH1 : histone H1
VIIL. Linked to ccg-9, pho-4 (658).
Cloned and sequenced.
Encodes histone H1(658).
hH2A : histone H2A
VII. Linked to Cen-VII (1T/18 asci), hH2B (on a common plasmid) (2032).
Cloned and partially sequenced: EMBL/GenBank AI392462; EST SC3B9.
Encodes histone H2A (2032).
hH2B : histone H2B
VII. Linked to Cen-VII and hH2A (on a common plasmid) (2032).
Cloned and sequenced: Swissprot H2B_NEUCR, PIR PN0142.
Encodes histone H2B (1032).
hH3 : histone H3
IIR. Adjoins hH4-1 (2248). On a common cosmid with hH4-1 and aro-9 (861). Between Fsr-55 and Fsr-3 (1447).
Cloned and sequenced: Swissprot H3_NEUCR, EMBL/GenBank X01612, GenBank NCHISH3, PIR S07350; EST NP4A11.
Encodes histone H3 (2248).
hH4-1 : histone H4-1
IIR. Adjoins hH3 (2248). On a common cosmid with hH3 and aro-9 (861). Between Fsr-55 and Fsr-3 (1447).
Cloned and sequenced: Swissprot H4_NEUCR, EMBL/GenBank X01611, GenBank NCHISH4.
Encodes histone H4-1 (2248).
hH4-2 : histone h4-2
IIIR. Linked to trp-1 (0/18 asci) (861).
Cloned and sequenced.
Encodes histone H4-2.
his : histidine
The biosynthetic pathway is shown in Fig. 38. Most histidine auxotrophs are inhibited by complex media or by certain combinations of amino acids with which histidine does not effectively compete for permeases of the basic, neutral, and general amino acid transport systems; see Fig. 47. A histidine mutant can grow on minimal medium plus histidine in the presence of either a basic amino acid or a competing neutral amino acid, but not in the presence of both (809, 1243, 1291). Histidine mutants were not obtained in early mutant hunts where complex media were used, but were recovered on histidine-supplemented minimal medium (809, 1159). General studies (329, 809, 2196). Enzymes of histidine biosynthesis are derepressed coordinately with those of tryptophan, arginine, lysine (292), and other amino acids [reviewed in ref. (1768)]. See cpc-1. Called hist.
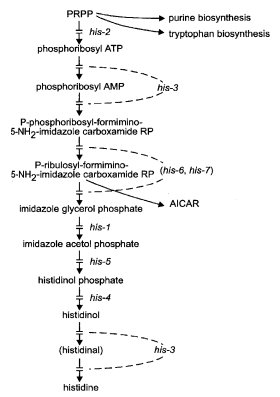
FIGURE 38 The biosynthetic pathway of histidine, showing the sites of gene action (23 , 42 , 43 , 329 , 1350 , 2196 ). Abbreviations: ATP, adenosine 5'-triphosphate; AMP, adenosine 5'-monophosphate; PRPP, 5-phosphoribosyl pyrophosphate. AICAR (5'-phosphoribosyl-5-aminoimidazole-4-carboxamide ribosyl phosphate) is an intermediate in purine synthesis. See Fig. 5. For relations between histidine and purine synthesis, see ref. (1543 ). Modified from ref. (1596 ), with permission from the American Society for Microbiology.
his-1 : histidine-1
VR. Between sp (11%), am (3%), ure-1 (1%) and pho-2 (3%), al-3, inl (1%; 10%) (218, 747, 809, 1086, 1122, 2014).
Requires histidine (809). Accumulates imidazole glycerol phosphate. Lacks imidazole glycerol phosphate dehydrase (EC 4.2.1.19) (42, 43) (Fig. 38). Expression appears positively regulated by a-isopropylmalate, a leucine biosynthetic intermediate that functions with leu-3 in leucine biosynthetic gene regulation (1048). Intralocus complementation (329). Recombination between his-1 alleles is controlled by rec-1 (314, 990, 2083). Called C84.
his-2 : histidine-2
IR. Between un-2 (<1%), T(AR190) and T(AR173)R, nuc-1 (l%) (314, 1331, 1580).
Cloned: pSV50 clones 10:6F, 6:11E, Orbach-Sachs clones X7H07, X20D08, X21G12, G13B04, G13H11, G19B11.
Requires histidine (809). Affects ATP phosphoribosylpyrophosphate pyrophosphorylase (EC 2.4.2.17) (23) (Fig. 38). Intralocus complementation (329). Recombination between his-2 alleles is controlled by rec-3 (337) and is not affected by rec-1 (314). Called C94.
his-3 : histidine-3
IR. Between met-10, lys-4 (1%) (1022, 1322) and cog (1%; 3%) (314, 338), ure-4 (1%) (156), ad-3A (1%; 3%) (492).
Cloned and sequenced: EMBL/GenBank AF045455, AF045456, M27531, EMBL NCHIS3, GenBank NEUHIS3; EST W08H2; pSV50 clones include 6:4E, 6:11C, 12:5C; Orbach-Sachs clone G3F06.
Requires histidine (809). Complex multidomain structural gene encoding histidinol dehydrogenase (EC 1.1.1.23), phosphoribosyl-ATP-pyrophosphohydrolase (EC 3.6.1.31), and phosphoribosyl-AMP cyclohydrolase (EC 3.5.4.19) (23, 1350) (Fig. 38). Mutants produce cross-reacting material (428). Supersuppressible amber alleles identified (303). Individual alleles may lack only the early reaction(s), or only histidinol dehydrogenase, or both. Mutants that lack only histidinol dehydrogenase accumulate histidinol (23, 329, 2196). Used to study intralocus complementation and recombination (22, 23, 45, 314, 329, 331, 336, 2194, 2195, 2197). Intralocus recombination is regulated by cog and rec-2 (45, 336) and is not affected by rec-1 (314). Translocation T(TM429), with one breakpoint in his-3, has been used to show that cog is cis-acting (336). Fine-structure interallelic complementation map (1511). Correlation of noncomplementing mutants with his-3 restriction site differences (557). Efficient integration of gene constructs by gene replacements at the his-3 locus (47, 1265, 1771). Differences in sequence between the Lindegren and St Lawrence strains (328). Called C140, T1710 (=C1710).
his-4 : histidine-4
IVR. Between cot-1 (1%; 4%) and met-5 (4%) (809, 1585).
Requires histidine (809). Lacks histidinol phosphate phosphatase (EC 3.1.3.15) (42, 43). Accumulates L-histidinol phosphate (Fig. 38). Allele P143h is heat-sensitive (wild type at 25ºC), whereas C141 is not heat-sensitive (1591); both are leaky (1412).
his-5 : histidine-5
IVR. Between pyr-3 (1%) and tol, trp-4 (3-7%) (329, 1466, 1911, 1928).
Requires histidine. Accumulates imidazole acetol phosphate and some imidazole glycerol phosphate. Evidently lacks imidazole acetol phosphate transaminase (EC 2.6.1.9) (23, 329, 2196) (Fig. 38). Intralocus complementation (329). Recombination control (314).
his-6 : histidine-6
VR. Between pyr-6 (6%; 18%), un-9 (6%), oak (6%) and T(NM149) (0/499), Tel-VR (30 kb) (1592, 1593, 1603, 1809).
Cloned (1809).
Requires histidine. Blocked prior to IGP (329, 2196), presumably in either EC 5.3.1.16 or EC 2.4.2.-, which catalyze the only two steps not definitively genetically characterized (Fig. 38). No intralocus complementation (95 alleles) (329). Intralocus recombination (314). Used in the first cloning of a Neurospora telomere (1809).
his-7 : histidine-7
IIIR. Between leu-1 (8%; 20%) and thi-2 (1% or 2%) (329, 426, 1582, 2044, 2196).
Cloned and partially sequenced: EMBL/GenBank AI398871; EST W09G5.
Requires histidine. Blocked prior to IGP, presumably in either EC 5.3.1.16 or EC 2.4.2.-, as the only two steps not definitively characterized genetically (329, 2196); likely encodes a bifunctional amidotransferase/cyclase, from EST (Fig. 38). Intragenic recombination (314).
hist : histidine
Symbol changed to his.
Histidine Sensitivity
Many mutagen-sensitive mutants are also sensitive to inhibition by histidine (see mus, uvs, mei, and Table 3) (946, 1467, 1830). Certain strains that were first identified as mutant on the basis of histidine sensitivity are not sensitive to the few mutagens tested [see, for example, ref. (520) and hss-1]. The mechanism underlying histidine sensitivity is not understood.
hlp-1 : histidinol permeability-1
VIIR. Between sfo (1%; 9%) and hlp-2 (8%; 25%), nt (28%; 37%) (879).
Enables a his-3 allele to use L-histidinol. Proposed to be due to increased uptake through basic L-amino acid transport system III [as defined in ref. (1522)]. The hlp-1 mutation confers increased sensitivity of lys and arg mutants to inhibition by arginine and lysine, respectively (879).
hlp-2 : histidinol permeability-2
VIIR. Between sfo (3%; 7%), hlp-1 (8%; 25%) and nt (29%) (879).
Enables a his-3 allele to grow on L-histidinol. Growth of his-3; hlp-2 on histidine is inhibited by methionine, isoleucine, valine, and asparagine (879).
hom : homoserine
IR. Between Tp(T54M94)R, arg-6 (1%) and al-1 (<1%), al-2 (2%; 7%), cnr (1%). Between breakpoints of T(STL76) and T(4637); hence, left of al-1 (1548, 1578, 1585). Located on a common cosmid with al-1 (1816).
Cloned and partially sequenced: EMBL/GenBank AI392256; EST SC2C6, pSV50 clone 3:11H.
Uses homoserine, or methionine + threonine (2066). Affects aspartate semialdehyde dehydrogenase (EC 1.2.1.11) (987) (Fig. 45). Inhibited on complex complete medium and by methionine and other amino acids (2066); supplemented minimal medium should therefore be used. Symbol changed from hs.
hph : hygromycin phosphotransferase
Introduced from Escherichia coli.
Neurospra crassa is sensitive to growth inhibition by hygromycin B. The bacterial gene hph encodes a phosphotransferase that confers resistance to the fungus (1977), where it has had numerous applications. The hph gene has been incorporated into cosmid vectors as a selectable marker for use in transformation (1502, 1504). Selection for hph expression has been used to obtain site-specific integration events (277). hph has been used to study gene silencing (955, 1529). As a reporter, hph has been used to obtain both cis- and trans-acting mutations that affect gene regulation by asking for increased hph expression when expression is normally repressed (677, 1241). Hygromycin phosphotransferase enzyme activity can be measured in N. crassa extracts (677, 678).
hpp : housekeeping phosphate permease
Between al-2 and nic-1 (1327).
Selected as a suppressor of the double mutant pho-4; pho-5, which lacks both of the two high-affinity permeases and is unable to grow at low phosphate concentration and high pH. On ordinary Vogel’s (high phosphate) medium, the triple mutant hpp; pho-4; pho-5 accumulates brownish pigment and does not grow as well as the pho-4; pho-5 double mutant (1327).
hs : homoserine
Symbol changed to hom.
hsp30 : heat shock protein 30
I. Linked to mei-3, Cen-I, un-2, his-2 (0T/18 asci) (1447).
Cloned and sequenced: Swissprot HS30_NEUCR, EMBL/GenBank M55672, J05601, EMBL NCHSP30, GenBank NEUHSP30, PIR A38360.
Encodes a 30-kDa heat-shock protein. Mutant obtained by RIP shows reduced viability at high temperature and altered location of a 22-kDa protein in mitochondria (1637). Lacks a small a-crystallin-related heat-shock protein, shows poor survival during heat shock on a nutrient medium with restricted glucose, and accumulates unphosphorylated glucose at high temperature. Hexokinase is reduced by more than 35% in the mutant relative to wild type. HSP30+ may protect hexokinase from thermal inactivation (1635).
hsp70 : heat shock protein 70
IIL. Between T(AR18)L and un-24, het-6, T(AR18)R (1935).
Cloned and sequenced: Swissprot HS70_NEUCR, EMBL/GenBank U10443, GenBank AAA82183.
Encodes a major stress-inducible 70-kDa heat-shock protein (1026), which reaches its highest level in late aerial hyphae (811). Homolog of Escherichia coli dnaK. A molecular chaperone involved in the translocation motor for the import of proteins through the mitochondrial membrane into the matrix (2300) (Fig. 64). Called hsps, hsps-1. For another member of the hsp70 family, see grp78.
hsp80 : heat shock protein 80
VR. Between inl (3T/18 asci), cya-2 (2T/18 asci) and tom70 (1 or 2T/17 asci). Linked to cmd, hsp83 (0T/18 asci) (1447).
Cloned and partially sequenced: EMBL/GenBank AI398579, PIR A56820; EST W01G6.
Encodes an 80-kDa heat-shock protein (1747); a cytosolic molecular chaperone of the eukaryotic stress 90 family (674). Hybridizes with Saccharomyces cerevisiae hsp83 probe (1636). Called hspe-1, hsp83.
hsp83 : heat shock protein 83
Allelic with hsp80.
hsp88 : heat shock protein 88
Unmapped.
Cloned and sequenced: Genbank AF069523, AAC23862.
Encodes an 88-kDa heat-shock protein that interacts with HSP30. Homolog of the mammalian Hsp110 family (1638).
hsp98 : heat shock protein 98
Unmapped.
Partially sequenced: Swissprot HS98_NEUCR, EMBL P31540, PIR S28174, Genbank AAB24758.
Encodes a 98-kDa heat-shock protein (2143) showing homology with HSP104 of Saccharomyces. HSP98 is enriched in the microsomal fraction of heat-shocked cells.
hspe-1 : heat shock protein 80
Symbol changed to hsp80.
hspp-1 : heat shock peroxidase
VI. Linked to Bml, Cen-VI, pan-2 (0T/18 asci) (1447).
Encodes a heat-shock-inducible peroxidase (1025).
hsps : heat shock protein 70
Symbol changed to hsp70.
hss-1 : histidine sensitive-1
IVR. Linked to cot-1 (19%), cys-4 (2%) (1593).
Sensitive to histidine (0.5 mg/ml) (1593), but not to UV (317), MMS, or γ rays (1828).
htb-1 : high-temperature banding-1
IVR. Linked to bd (27%), near T(R2355), cot-1 (1433).
Allows clear expression of circadian conidiation rhythm at 36ºC (1433).
htb-2 : high-temperature banding-2
Unmapped. Unlinked to bd (1433).
Allows clear expression of circadian conidiation rhythm at 36ºC (1433).
htl : hyphal tip lysis
IR. Right of ad-3 (1%; 10%) (204). Linked to nuo12.3 (3/18 asci), cys-9 (1T/18 asci) (1447).
Cloned: Orbach-Sachs clone G8H12 (204).
Encodes an abundant 19-kDa protein of unknown function. Mutants generated by RIP show lysis of hyphal tips when grown on solid medium. Patches of bright orange-red pigment are accumulated. Hyphal tips below the agar surface produce large balloons or lens-shaped structures. Growth on high concentrations of NaCl is indistinguishable from that of wild type (204). Called vac-5 (1447).
Return to the 2000 Neurospora compendium main page