Lactose Utilization
Lactose is a poor carbon source for Neurospora (127). The ability to use lactose has a multigenic basis. In early studies, strains were identified that showed impaired ability to grow on lactose. Properties of b- D-galactosidase (lactase) were unaltered; levels were normal when grown on sucrose, but depressed on lactose (1136, 1137). These strains differed from wild type at several loci, each with a small and additive effect on lactose utilization; e.g., three component genes from “lac” strain 31389 ´ wild type were shown to be unlinked and were designated n-lac-1, pow(n-lac-2) (powdery conidia), and floc(n-lac-3) (flocculent morphology). These genes are not specific for lactose utilization, however; they also showed an altered adaptation response to other carbon sources. No major gene was identified that qualifies to be designated lac (665). The failure to find a single-gene mutant can be ascribed to the fact that Neurospora has two b-galactosidases (127, 1179).
lacZ : lactose Z
From Escherichia coli.
The E. coli structural gene for b-galactosidase is a reliable reporter gene for quantitative measures of gene expression in Neurospora crassa (574, 1771). For examples, see refs. (80), (146), (409), (1237), (1238), (1517), and (2209). Enzyme activity from the introduced lacZ is manyfold higher than that of the endogenous Neurospora b-galactosidases (1771).
lacc : laccase
VIIL. Between nic-3 (14%) and thi-3 (7%) (2296).
Cloned and sequenced: Swissprot LAC1_NEUCR, LAC2_NEUCR, EMBL/GenBank M18333, M18334, EMBL NCLCCA, NCLCCB, GenBank NEULCCA, NEULCCB.
Laccase structural gene (EC 1.10.3.2) (benzenediol:oxygen oxidoreductase, urishiol oxidase), extracellular. Alleles from wild-type strains TS and OR sequenced (726). The laccase polypeptide is processed posttranslationally at both its N- and C-termini (726). lacc mutants do not show a detectable phenotype except that laccase is not produced (2296). Gene expression is induced by inhibitors of RNA or protein synthesis or by certain aromatic molecules (684, 1209). The secreted enzyme forms a blue halo of oxidized o-tolidine around colonies in test plates. Screening for mutants that overproduced laccase led to the identification of lah-1 (2041). Screening for additional mutants that did not allow the overexpression of laccase in the lah-1 background established that functional cpc-1 was required for lacc induction (833, 2042, 2296). Called lni-2.
lah-1 : laccase halo-1
IL. Between nit-2 (2%) and leu-3 (5%) (2041).
Laccase is derepressed. Hypersensitive to cycloheximide (2041). Acts as a negative regulator of cpc-1 (833). Identified by the development of a blue halo when o-tolidine is added to colonies of exo-1 grown on xylose plus sorbose (2041). Used to obtain a mutant in lacc, the laccase structural gene (2296). Called halo-1 (832).
lao : L-amino acid oxidase
IIIR. Linked to un-17 (5%) (511)
Cloned and sequenced: Swissprot OXLA_NEUCR, EMBL J05621, PIR A38314 (1480).
Structural gene for L-amino acid oxidase (EC 1.4.3.2) (1480). Under general nitrogen metabolite regulation (1914) (Fig. 50). Also called lox.
le-1 : lethal-1
IVR. Linked to pan-1 (1% or 2%) (719). Right of cot-1 (14%) (1405, 1408).
Ascospores containing le-1 are black, but they fail to germinate unless given special treatment (719). Autonomously expressed. Growth is colonial and aconidiate, with dense granular aerial mycelium turning brown with age (1405, 1408). Photographs (719, 1408). Cell-wall peptides are reduced in amount (2249). Alleles B55 and S4355 of ref. (719) were presumed to be allelic with similar mutants called col-le-1 (CM3) and col-le-2 in ref. (1405) and (1408), but direct tests were not made.
le-2 : lethal-2
VIIL. Linked to met-7 (7%). Indicated to the left (719).
Ascospores containing le-2 are black, but they fail to germinate except for a few that are recovered after aging. Autonomously expressed. Compact colonial growth (719).
leu : leucine
For the biosynthetic pathway, see Fig. 39. Leucine mutants have been used extensively for studies of regulation (799, 1281, 1640). When grown on Difco Bacto Agar-sorbose medium, leucine auxotrophs acquire suppressors that are leaky auxotrophs blocked at various steps in sulfur metabolism. Apparently, these blocks allow more efficient use of the traces of leucine in the agar (793, 797). Most aliphatic and aromatic amino acids can inhibit the growth of leucine mutants at appropriate concentrations, probably as a result of competition for a common uptake system (793).
leu-1 : leucine-1
IIIR. Between ad-4 (1%; 5%), col-6 (1%) and his-7 (8%) (426, 985, 1122, 1591, 2044)
Cloned and sequenced: Swissprot LEU3_NEUCR, EMBL/GenBank U01061, EMBL NC061; EST NP2D4.
Requires leucine (1705, 1706). Structural gene for a-isopropylmalate dehydrogenase (EC 1.1.1.85) (798) (Fig. 39). Accumulates a-isopropylmalate (a-IPM) and b-isopropylmalate (799). Synthesis of enzyme also requires the function of regulatory gene leu-3+ and the presence of a-IPM, which acts as an inducer (799). Resistant to aminotriazole (1048). Female sterile (1424). Used to study reversion and competition in heterokaryons (1762). The 5'-leader of the mRNA precursor contains an intron that does not interrupt a coding region (1190). Allele 33757 contains a 1-bp insertion predicted to result in a frameshifted, prematurely terminated polypeptide (1190).
leu-2 : leucine-2
IVR. Between trp-4 (2%) and ilv-3 (4%) (1124, 1928).
Cloned (1047).
Requires leucine (1705, 1706). Structural gene for isopropylmalate isomerase (EC 4.2.1.33) (804, 1709) (Fig. 39). Altered heat inactivation of hybrid enzymes (804). Structural differences of hybrid enzymes (1709). Accumulates a-IPM (799). Synthesis of enzyme also requires the function of regulatory gene leu-3+ and the presence of a-IPM, which acts as an inducer (799). Induction measured at the transcript level is very rapid (1276). Resistant to 3-aminotriazole (1048). Alleles show intralocus complementation (796). Allele 37501 is heat-sensitive (30 vs 20ºC), leaky at 25ºC (1704).
leu-3 : leucine-3
IL. Between nit-2 (12%; 18%), lah-1, In(OY323)L and T(OY322)L, cyt-1 (5%; 8%), T(OY321) (115, 1578, 1582).
A regulatory gene. The mutant requires leucine (1705). Prevents the synthesis of a-isopropylmalate isomerase and b-isopropylmalate dehydrogenase and prevents full de-repression of a-isopropylmalate synthetase. Is also involved in the regulation of isoleucine and valine synthesis (799, 1497, 1640) (Fig. 39). The LEU-3 regulator, which from the rapid rate of LEU-3-dependent leu-2 induction appears to be present irrespective of inducing conditions (1276), may also have more global regulatory functions (52). Neurospora crassa LEU-3 might be a Zn(II)2Cys6 binuclear cluster DNA-binding protein, as is its functional yeast homolog, LEU3 (2094). However, the DNA-binding sequences used by the Saccharomyces cerevisiae protein are not found in the promoter region that is upstream of the Neurospora leu-1 mRNA transcript (1190), which is regulated by LEU-3. The original mutant allele, 47313, is leaky, but some other alleles, e.g., R156, are not.
leu-4 : leucine-4
IL. Between cyt-1, T(OY321) and cys-5 (<<1%) (801, 1578, 2198).
Cloned: pSV50 clone 9:7A.
Requires leucine. Structural gene for a-isopropylmalate synthetase (EC 4.1.3.12) (798, 799, 804) (Fig. 39). Feedback-negative mutants (798, 799). Hybrid synthetases with altered properties (804). Complementation between alleles (804, 2198).
leu-5 : leucine-5
VR. Between cyh-2 (1%) and sp (3%; 9%) (378, 1582, 1603, 1651).
Cloned and sequenced: Swissprot SYLM_NEUCR, EMBL/GenBank M30472, PIR A33474, SYNCLM, EMBL NCLEURS, GenBank NEUMTLEURS.
Structural gene for mitochondrial leucyl-tRNA synthetase (EC 6.1.1.4) (378, 1651). Mutant 45208 has a partial leucine requirement at low temperatures, a tighter leucine requirement at 34ºC, and stops growth at 37-39ºC, regardless of leucine supplementation (556, 1651). The 45208 allele contains a Thr to Pro mutation at amino acid 135, which is presumed to result in the phenotypic defect (378). Mutations mapping in the leu-5 region appeared to affect either cytoplasmic leucyl-tRNA synthetase or mitochondrial leucyl-tRNA synthetase separately or both simultaneously (138, 803), and leu-5 was proposed to be a gene complex consisting of structural genes for both enzymes. This proved to be incorrect; the cytoplasmic leucyl-tRNA synthetase is specified instead by leu-6 (153). In mitochondrial mutant [cni-3], mitochondrial tRNA synthetase is increased greatly whereas the cytoplasmic enzyme is unchanged (802). Allele 45208 is somewhat unstable (803). It causes alterations in unrelated enzymes, apparently via mistranslation (1183, 1651), and it was used to study the hypothesis that senescence is due to faulty protein synthesis (1183). Assembly of glycerol kinase and GPDH into the inner mitochondrial membrane is not impaired in leu-5 (421). Recovery from ascospores is poor at 34ºC (on complex complete medium); ascospores are best germinated at 25ºC.
leu-6 : leucine-6
IIR. Between trp-3 (2/16), Fsr17 (0/16; 5T/18 asci) and Tel-IIR (1T, 1NPD/16 asci) (153, 378, 1447).
Cloned and sequenced: Swissprot SYLC_NEUCR, EMBL/Genbank, M30473, X13021, PIR A33475, S04532, SYNCLC, EMBL NCLEUR01, NCLEURSC, GenBank NEULEURSC; EST SM1D7.
Structural gene for cytoplasmic leucyl-tRNA synthetase (EC 6.1.1.4) (153, 379, 1113). The reported effect of leu-5 mutations to alter the Km of the cytoplasmic leucyl-tRNA synthetase in crude extracts (138) was not reproduced with the purified cytoplasmic enzyme (1113), although the stability of the enzyme in extracts was decreased when extracts were prepared from cells incubated at nonpermissive temperatures. Subject to cross-pathway control through cpc-1. Histidine starvation induces new transcription start sites (379).
lgd : laggard
VIL. Linked to chol-2 (7/32). Left of T(OY350) (1589).
Growth is delayed greatly following ascospore germination. Morphologically wild type when grown. Obtained in progeny of Dp(VIL®IR)OY350 ´ Normal, presumably by RIP (1589).
Lipid Biosynthesis
See Fig. 18.
lis-1 : light insensitive-l
IR. Between ad-3 (6%) and al-1 (16%) (1514).
Circadian conidiation is not suppressed in constant light (1514). The growth rate is normal. Photoinduced genesis of carotenoids and phase shifting of periodic conidiation are not altered (1711). Recessive in heterokaryons.
lis-2 : light insensitive-2
VI. Between chol-2 (11%) and trp-2 (26%) (1515).
Resembles lis-1, but the growth rate is reduced somewhat (1515, 1711).
lis-3 : light insensitive-3
VR. Right of inl (4%) (1514).
Resembles lis-1, but the growth rate is reduced markedly(1514, 1711).
lni-1 : laccase noninducible-1
Allelic with cpc-1.
lni-2 : laccase noninducible-2
Changed to lacc-1.
lox : L-amino acid oxidase
Symbol changed to lao.
lp : lump
II. Right of thr-3 (10%). Linked to bal (25%) (1585, 1603).
Restricted colonial growth. Grows more rapidly then bal and forms aerial hyphae (1585).
lpl : lysophospholipase
IR. Right of his-3 and cog, which it adjoins (2276).
Cloned and sequenced: EMBL/GenBank AF045574, AF045575.
Encodes lysophospholipase (EC 3.1.1.5). Shows homology to Saccharomyces PBL1 and SPO1. Alleles differ in wild types St. Lawrence 74A and Lindegren 25a (2276).
lys : lysine
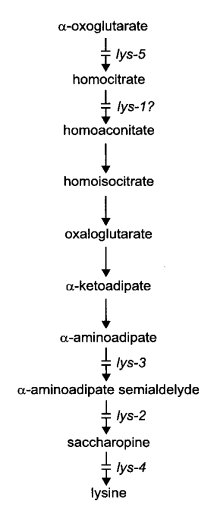
Lysine biosynthesis is by the a-aminoadipate pathway in Neurospora and other higher fungi ( 2163) (Fig. 40). Enzymes of lysine biosynthesis are subject to cross-pathway control (1768); see cpc. All lysine auxotrophs are inhibited competitively by arginine (548). (See the arg entry for a medium that allows growth while providing both lysine and arginine requirements.) Resistance to arginine is conferred on lys-1 by a presumed transport mutant argR. Complex interactions between lys, pyr, and arg mutants have been described (910).

FIGURE 40 The biosynthetic pathway of lysine, showing sites or probable sites of gene action (246 , 751 , 887 , 933 , 1479 , 2113 ). alpha -amino-e-hydroxycaproic acid can be converted to alpha -aminoadipate semialdehyde (2292 ), but apparently it is not an intermediate. Modified from ref. (1596 ), with permission from the American Society for Microbiology.
lys-1 : lysine-1
VL. Between caf-1 (4%; 14%) and cyt-9 (5%), at (1%; 20%) (926, 1582, 1598).
Uses lysine, a-aminoadipic acid, or e-hydroxynorleucine (a-amino-e-hydroxycaproic acid) (751, 2113). Accumulates homocitrate on limiting lysine concentrations (887) (Fig. 40). Fine structure; complementation between alleles (15).
lys-2 : lysine-2
VR. Between ilv-1, ilv-2 (4%; 7%) and cyh-2 (<1%), leu-5 (9%) (10, 1603, 1651).
Requires lysine. Will not use e-hydroxynorleucine (751). Probably blocked in α-amino acid reductase
(EC 1.2.1.31), converting aminoadipic semialdehyde to saccharopine (2113) (Fig. 40).
lys-3 : lysine-3
IR. Between al-1 (9%) and In(OY323)R, ace-3, nic-1 (9, 1971). Not included in duplications from T(OY323) ´ In(NM176); hence, left of ace-3 (115, 1578).
Cloned by complementation (1815)and sequenced: GenBank 142777.
Requires lysine. Partial response to glutaric acid (1478). Lacks homocitrate synthase activity (EC 4.1.3.21) (887, 1478) (Fig. 40). Some alleles, such as 37402, are autonomous ascospore lethals or semilethals, producing mostly immature white spores [photograph in ref. (1986)]. Viability of the ascospores carrying the mutant allele is improved by long incubation (14). With other alleles, such as DS6-85, ascospores blacken and germinate normally. Malate and citrate are accumulated on limiting lysine concentrations (887). Four complementation groups have been identified (19). Allele 37402 was called asco.
lys-4 : lysine-4
IR. Between nuc-1 (1%), met-10 and his-3 (1%) (492, 1022, 1331).
Requires lysine (751). Lacks saccharopine-cleaving enzyme activity, saccharopine dehydrogenase (L-lysine-forming) (EC 1.5.1.7) (2113) (Fig. 40). Complementation between alleles (20). Uses 0.5 mg/ml lysine.
lys-5 : lysine-5
VIL. Between cyt-2 (6%), aro-6 (3%), and un-4 (2%; on a common cosmid) (1596, 1815, 1986).
Cloned by complementation (1815).
Requires lysine. Partial response to glutaric acid (1478). Lacks homocitrate synthase activity (EC 4.1.3.21) (887, 1478) (Fig. 40). Some alleles, such as 37402, are autonomous ascospore lethals or semilethals, producing mostly immature white spores [photograph in ref. (1986)]. Viability of the ascospores carrying the mutant allele is improved by long incubation (14). With other alleles, such as DS6-85, ascospores blacken and germinate normally. Malate and citrate are accumulated on limiting lysine concentrations (887). Four complementation groups have been identified (19). Allele 37402 was called asco.
lysR : lysine resistant
IR. Between his-3 and nic-2 (878, 1083).
Growth of arg-1 lysR is resistant to normal inhibition by L-lysine. Proposed to be due to basic amino acid transport system (1083). May be allelic with su(mtr)-1 (1082).
Return to the 2000 Neurospora compendium main page