m : microconidial
Used for pe.
ma-1 : malate utilization-1
Unmapped. Probably in a left arm, any of III-VII.
Unable to use malate as the carbon source when the tricarboxylic acid cycle is blocked by a suc (pyruvate carboxylase) mutation. Altered mitochondrial malate dehydrogenase. Scorable only in suc (1393, 1396, 1398).
ma-2 : malate utilization-2
IIR. Between un-20 and ace-1 (210)
Unable to use malate as the carbon source when the tricarboxylic acid cycle is blocked by a suc (pyruvate carboxylase) mutation. Altered mitochondrial malate dehydrogenase. Scorable only in suc (1393, 1396, 1398).
mac : methionine-adenine-cystine
Used for met-6 allele 65108. See met-6.
mad-1 : MADS box-1
I. By correlation with cosmids assigned to linkage groups (573).
Cloned: Orbach-Sachs clone G11C02 (1268).
Cloned by PCR with degenerate primers encoding MADS box conserved domains. The predicted protein is virtually identical to the product of Saccharomyces MCM1 (1268). Proteins including MADS box domains are DNA-binding proteins present in plants, animals, and fungi. The Mcm1 protein is a regulator of mating-type specific gene expression in Saccharomyces (1268).
mak-1 : mitogen-activated protein kinase-1
Unmapped.
Cloned: Orbach-Sachs clone X13B11 (1267).
Cloned by PCR with degenerate primers encoding MAPK (mitogen-activated protein kinase)-conserved domains (1267). The predicted protein product shows similarity to the product of Saccharomyces MPK1 (also known as SLT2) (1267).
mak-2 : mitogen-activated protein kinase-2
VII. By correlation with cosmids assigned to linkage groups (573).
Cloned: Orbach-Sachs clone G13A03 (1267).
Cloned by PCR with degenerate primers encoding MAPK (mitogen-activated protein kinase)-conserved domains (1267). The predicted protein shows similarity to the products of Saccharomyces FUS3 and KSS1, which are involved in mating pheromone response in budding yeast (1267).
mak-3 : mitogen-activated protein kinase-3
Unmapped.
Cloned and partially sequenced: EMBL/GenBank AI398492, AI416420; EST W08A7, W08A8.
The predicted protein resembles the product of HOG1 of Saccharomyces, which is involved in cell-wall regulation and response to osmotic stress. Also resembles HOG1 of Zygosaccharomyces rouxii (1267).
mat : mating type
IL. Between un-3 (0.1% or less) and un-16 (<1%) (917, 1470, 1546). Between eat-5 and eat-1 (1691).
Genes at the mat locus are master regulators of mating, postfertilization development, and nuclear identity. Strains must be of opposite mating type, mat A and mat a, for a complex of events to occur that are associated with sexual reproduction and morphogenesis. These include the attraction of trichogynes to cells of opposite mating type (82, 182), pickup and transport of nuclei to the ascogonium, growth and development of perithecia, proliferation of heterokaryotic ascogenous hyphae, conjugate nuclear divisions in precrozier and crozier cells, and karyogamy. Vegetative cultures deleted for the mat genes are viable, grow normally, and form protoperithecia (629).
In Neurospora crassa, strains of opposite mating type are vegetatively incompatible. mat A + mat a combinations are unable to form stable heterokaryons (134, 721, 1629, 1786). Vegetative fusion is usually followed by cell death (721), but some A + a heterokaryons grow slowly (517, 779, 794). Heterozygous A/a duplications are highly abnormal, with inhibited growth, spider-like morphology, and darkening of agar (1475, 1561). Incompatibility in heterokaryons or in heterozygous duplications is relieved by the spontaneous deletion of either allele (517, 1468). Vegetative incompatibility is not expressed during the sexual phase. An active allele at the unlinked tol locus is necessary for the mat-mediated incompatibility reaction to occur in either heterokaryons or duplications. tol does not affect sexual compatibility or mating, however (1466). The sexual and vegetative manifestations of mat have been resolved mutationally, but not by genetic recombination (1470). Mutants selected by loss of vegetative incompatibility usually lose both sexual and vegetative functions simultaneously, and both functions usually are restored simultaneously in revertants selected for restoration of fertility (one null mutant gives atypical revertants) (517, 777, 779).
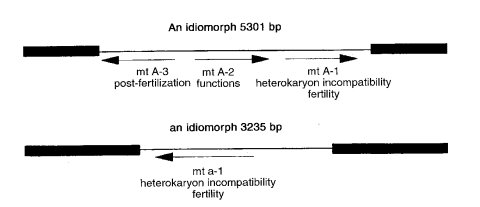
The mat locus is occupied by nonhomologous genes in the two mating types. mat A and mat a therefore are called idiomorphs rather than alleles (1332). Both idiomorphs have been cloned and sequenced (see following entries). The A idiomorph contains three open reading frames, the a idiomorph only one (Fig. 41). mat A-1 and mat a-1 appear to be essential for both mating and sexual development, whereas mat A-2 and mat A-3 increase the efficiency of sexual development but are not essential for ascospore production (629). When the idiomorph in a mat A strain is replaced by the a idiomorph, the strain becomes fully functional as mat a (356). All target genes essential for development as mat a therefore are present in strains of the opposite mating type.
Classical genetics has failed to reveal the presence of any genes related to mating functions in regions flanking mat, but molecular analysis has revealed regions contiguous to mat that are involved in mating and sexual development. A flanking region right of mat is divergent in sequence between species and often between mating types of the same species (1337, 1690). This region contains eat genes that appear to be involved in mating. eat-1 is transcribed in mat-A strains but not in mat-a, in which a smaller transcript is produced. An allele of eat-2 that has been inactivated by RIP is recessive vegetatively (slow growing, aconidiate), but is dominant (sterile) in the sexual phase (1691). Genes linked to mating type encode putative pheromone precursors that are expressed specifically in mat A (ccg-4) or in mat a (mfa-1); see also scp.
In the early literature, A was called + (plus) or A, whereas a was called – (minus) or B [e.g. ref. (547)]. The locus symbol was changed from mt to mat in 1997 to conform with usage in most other fungi (745). For reviews of mating type, including homologies and comparisons with other species and genera, see refs. (137), (407), (744), (1098), (1639), and (1976). Reference (407) is concerned especially with sexual development.

FIGURE 41Structural and functional regions of the mating type idiomorphs of N. crassa. The idiomorphic DNA sequences are indicated by lines, and the conserved flanking regions are indicated by boxes. Arrows indicate the direction of transcription. From ref. (744 ) [Glass, N. L., and M. A. Nelson (1994). Mating-type genes in mycelial ascomycetes. In "The Mycota" (R. Brambl and G. A. Marzluf, eds.), Vol. I, pp. 295-306, Fig. 1.], with permission. Copyright 1994 Springer-Verlag.
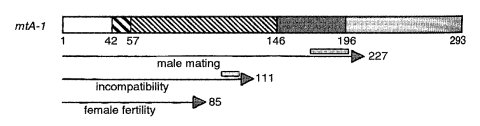
FIGURE 42 Summary of the proposed regional specialization in the mat A-1 polypeptide of N. crassa, which is symbolized by a rectangle. The finely hatched rectangle encompasses a region of similarity to FMR1 of Podospora, MAT-1 of Cochliobolus, MAT 1of Kluyveromyces, and MAT 1p of Saccharomyces. The coarsely hatched area (positions 42-57) corresponds to the region of greatest similarity to MAT 1p of Saccharomyces. The dark-shaded area represents the region of similarity unique to MAT A-1 and FMR1 of Podospora. The lightly shaded area represents the acidic C-terminus. The arrows represent the regions found to be minimally sufficient for the functions noted. The shaded segments above the arrows represent regions found to be required for the activity noted. From ref. (1796 ) [Saupe, S., L. Stenburg, K. T. Shiu, A. J. Griffiths, and N. L. Glass (1996). The molecular nature of mutations in the mt A-1 gene of the Neurospora crassa A idiomorph and their relation to mating-type function. Mol. Gen. Genet. 250: 115-122, Fig. 5.], with permission. Copyright 1996 Springer-Verlag.
mat A : mating type A
IL.
Cloned and sequenced: Sequences for component genes follow; Orbach-Sachs clones X1B02, X5A06, X5E06, X5E07, X11D06.
Symbol for the A idiomorph, with three open reading frames, mat A-1, mat A-2, and mat A-3, which are read divergently (630, 631) (Fig. 41). The abbreviated symbol A or mat A is preferred for use in contexts where the A-1, A-2, and A-3 constitution is irrelevant.
mat A-1 : mating type A-1
IL. Right of mat A-2.
Cloned and sequenced: Swissprot MATA_NEUCR, EMBL/GenBank M33876, PIR S65582, EMBL NCAMTR, Genbank NEUAMTR.
Encodes a DNA-binding protein similar to Podospora FMR1 in mating function (53). Mutational analysis (1796) (Fig. 42). The MAT A-1 polypeptide has a short but decisive stretch of homology with MAT1p of Saccharomyces. Inactivation by RIP shows mat A-1 to be essential for mating identity, mating functions that follow fertilization, and mat-associated vegetative incompatibility (743, 1796). Mutant mat A-1m99, which is heterokaryon-compatible and fertile with mat a when used as a female but not as a male, is due to a base-pair substitution that results in a truncated protein. Mutant A-1m13, which is heterokaryon-compatible and produces perithecia but no ascospores with mat a, is a frameshift (1796). Other mutants lose both the mating and the vegetative incompatibility functions simultaneously (777) as a result of frameshifts in mat A-1 (741).
mat A-2 : mating type A-2
IL. Between mat A-1 and mat A-3.
Cloned and sequenced: Swissprot MATC_NEUCR, EMBL/GenBank M33876, PIR S65583, EMBL NCAMTR, Genbank NEUAMTR.
Encodes a putative DNA-binding protein similar to Podospora SMR1 in mating function (631). Strains mutant in mat A-2 are normal in mating and fertility but are unaffected in vegetative incompatibility (629, 743). Strains mutant in both A-2 and A-3 are able to mate normally but are impaired for postmating steps in ascospore genesis, suggesting that MAT A-2 and MAT A-3 are redundant (629).
mat A-3 : mating type A-3
IL. Left of mat A-2.
Cloned and sequenced: Swissprot MATD_NEUCR, EMBL/GenBank M33876, PIR S65584, EMBL NCAMTR, Genbank NEUAMTR.
Encodes an HMG box DNA-binding protein similar to Podospora SMR2 in mating function (631, 1615). MAT A-3 contains a transcription activation domain (83). Not concerned with vegetative incompatibility. Strains mutant in A-3 are normal in mating and fertility. Strains mutant in both A-2 and A-3 are also able to mate but are impaired for postmating steps in ascospore genesis, suggesting that MAT A-2 and MAT A-3 are redundant (629).
mat a : mating type a
IL.
Symbol for the a idiomorph, containing a single open reading frame (356). The abbreviated symbols a and mat a are preferred in most contexts.
mat a-1 : mating type a-1
IL.
Cloned and sequenced: Swissprot MATB_NEUCR, EMBL/GenBank M54787, Genbank NEUMTA1A, Orbach-Sachs clones X13E01, X22F11 (1978).
Encodes an HMG box DNA-binding protein, presumed to be a transcriptional regulator. Similar to Podospora FPR1 in mating function (53). The HMB box region is sufficient for DNA binding but not for mating-type activity. DNA-binding activity does not appear to be necessary for vegetative incompatibility (1614), but does appear to be necessary for mating activity. Changes within the carboxy-terminal region of the polypeptide eliminate vegetative incompatibility function without affecting mating-type function. The mat a gene is essential for mating identity, mating functions after fertilization, and mat-associated vegetative incompatibility (1614). Mutant am33, which is both heterokaryon-compatible and sexually competent with mat A strains, is due to a single-nucleotide change that alters the protein product near the carboxy terminus (1978). Other known mat a-1m mutants have simultaneously lost both mating ability and vegetative incompatibility (777, 779).
Mating Type
Symbol changed from mt to mat. See mat.
mb-1 : male barren-1
VII. Linked to nic-3, wc-1 (23%) (1582, 2156).
Perithecial development is blocked when mb-1 is used as male parent in crosses to an mb+ female. Many perithecia are produced. These are mostly small, brown, and without beaks or ascospores, but a few become mature and produce ascospores (2156). Perithecia are normal and fertile when mb-1 is used as female parent, fertilized by mb+ (2202). Events following pachytene are not completely normal in the female, however, as observed cytologically (1673). Homozygous barren (1582). Recessive in heterokaryons, complementing mb-2 and mb-3 (2157). One occurrence, allele 8455.
mb-2 : male barren-2
IR. Between cyh-1 (5%) and al-1 (7%) (1582).
Perithecial development is blocked when mb-2 is used as male parent to fertilize an mb+ female. Many perithecia are produced, mostly small and brown, without beaks or ascospores; a few perithecia mature and produce ascospores (2156). Perithecia are normal and fully fertile when mb-2 is used as female parent and fertilized by mb+ (2202). Homozygous barren (1582). Recessive in heterokaryons, complementing mb-1 and mb-3 (2157).
mb-3 : male barren-3
IR. Linked to cyh-1 (18%), al-1 (2%), mb-2 (6%) (1172, 1582).
Perithecial development is blocked when mb-3 is used as male parent to fertilize an mb+ female. Many perithecia are produced, mostly small, brown, and without beaks or ascospores; a few perithecia mature and produce ascospores (2156). Perithecia are normal and fully fertile when mb-3 is used as female parent and fertilized by mb+ (2202). Development of perithecia may then be slower than normal, however (1673). Homozygous barren (1582). Recessive in heterokaryons, complementing mb-1 and mb-2 (2157). Six occurrences.
mcb : microcycle blastoconidiation
VR. Linked to al-3 (3%) (1250), cyh-2, sod-2 (0T/18 asci) (1447).
Cloned and sequenced: EMBL/GenBank L78009, EMBL NCMCB, GenBank NEUMCB; Orbach-Sachs clone X13C10.
Encodes the regulatory subunit of cyclic AMP-dependent protein kinase (PKA). The mutant has increased PKA activity. The gene consists of two overlapping transcriptional units. Discovered as a mutant allele extracted from a wild-collected strain (1250). Cultures grown on agar are morphologically wild type at 25º-30ºC. When macroconidia from agar-grown mycelial cultures are germinated in liquid shake culture at 25ºC, they generate hyphae that consist of chains of spherical cellular compartments (1250) (Fig. 43). Similar morphology is observed when hyphae grown at 25ºC are switched to 37ºC. When conidia are germinated at 37ºC, the location of septa is abnormal, actin is disorganized, and conidia grow into spheres that enlarge and ultimately burst (247). Apolar growth increases the secretion of extracellular proteins (1152). Apolar growth is suppressed by cr-1. mcb may therefore provide an opportunity to select for new forward mutations at loci involved in the cAMP-dependent kinase regulatory pathway (1632). Intracellular actin levels do not increase in the mcb mutant under conditions that substantially increase the amount of growing surface area (2092).
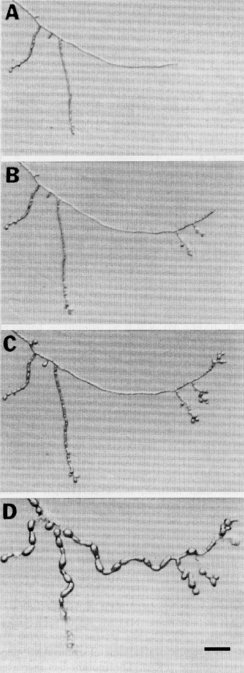
FIGURE 43 Loss of growth polarity in the mcb mutant at restrictive temperature. (A) Hyphae of an mcb strain after overnight incubation at 25C on a dialysis membrane overlying sucrose minimal agar medium. The hyphae shown in (A) were shifted to 37C and photographed after 1.5 (B), 3 (C), and 9hr (D). Note that growth polarity is lost in all regions of mcb hyphae grown at 37C for 9 hr, and that apolar growth continues until hyphal compartments burst (D). Bar length = 100 mm. From ref. (247 ) [Bruno, K. S. et al. (1996). EMBO J. 15: 5772-5782.], with permission from Oxford University Press.
mcm : microcycle microconidiation
IIL. Linked to ro-7 (1%) (1250, 1251).
When macroconidia from agar-grown mycelial cultures are germinated in liquid shake culture at 22ºC, the germlings produce uninucleate microconidia within 24 hr, reaching concentrations of 107 /ml at 72 hr (1251). At 30ºC, multinucleate arthroconidia and few or no microconidia are produced. The morphology of surface-grown cultures is normal wild type (Fig. 44).
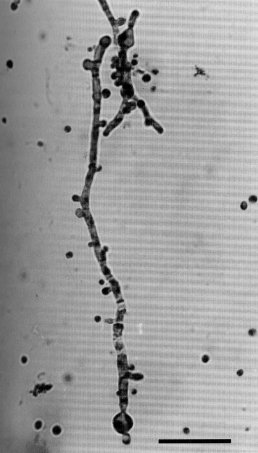
FIGURE 44 Microcycle microconidiation by an mcm strain ~12 hr after inoculation of air-grown macroconidia into liquid shake culture at 22C. Microconidia are produced as lateral protuberances (arrows) on the microcycle structure produced by a germinated macroconidium. Free microconidia also are present. Bar length = 25 mm. From ref. (1250 ), with permission from the Society for General Microbiology.
md : mad
VR. Between sh (3%) and sp (9%) (569).
Spreading growth with a characteristic branching pattern. Modifies the banding phenotype of cl. Photographs (569).
mdk-1 : mitotic division kinase-1
Unmapped.
Cloned by degenerate primers encoding MAPK (mitogen-activated protein kinase)-conserved domains (1267).
The predicted protein is very similar to the product of Saccharomyces PHO85, a cdc2-like kinase (1267). Characterization of genes regulating phosphorus uptake and metabolism in Neurospora predicts that the product of the uncloned pgov gene will be functionally and structurally like Pho85p (1322), suggesting that mdk-1 and pgov may be allelic. Attempts to recover the gene from the Orbach-Sachs cosmid library or to clone it from the Neurospora chromosome have failed (1267), as has an attempt to retrieve it by tagged mutagenesis (1322). This suggests that the gene may be toxic in Escherichia coli.
mdk-2 : mitotic division kinase-2
III or IV. By correlation with cosmids assigned to linkage groups (573).
Cloned: Orbach-Sachs clones G15B07, G15D07 (1267).
Cloned by PCR with degenerate primers encoding MAPK (mitogen-activated protein kinase)-conserved domains (1267). The predicted protein shows similarity to the product of S. pombe dsk1 (“division suppressor kinase”), which was cloned in S. pombe as a multicopy suppressor of another division kinase. The mutant gene shows no obvious phenotype in S. pombe, however. No homologue is present in the Saccharomyces genome (1267).
me : methionine
Symbol changed to met.
mea-1 : methylammonium resistant
I. Right of acr-3 (9%) (2220).
Resistant to inhibition by methylammonium, a structural analog of ammonium. nit-2; mea-1 double mutants show nitrogen-starved growth on ammonium (566). Confers resistance to 50 mM methylamine in nitrate medium (2220).
med : medusa
IVR. Linked to met-5 (5%), pan-1 (8%) (719).
A slow-growing, spreading morphological mutant forming distinctive grooves on the surface of agar (719). Photograph (719). Cell-wall peptides are reduced in amount (2249).
mei : meiotic
This name has typically been used for mutants having partial or almost complete blocks to meiosis and ascus development. Both recessive and dominant meiotic mutants are known. Crosses homozygous for recessive mutants usually produce few or no viable black ascospores; those that germinate may show much reduced meiotic recombination and greatly increased nondisjunction. Abnormal segregation results in white, inviable, and hypohaploid ascospores. Production of white ascospores provides a basis for isolation and scoring (1933). Sexual-phase recessive mutants that affect meiotic development are very common in natural populations of Neurospora crassa (1174). Some of the mutants designated mei specifically affect meiotic or premeiotic events (e.g., mei-1 and mei-4), and similar mutants may have been given other names such as asc, fmf-1, or mb. Other meiotic-defective mutants may be more generally deficient in recombinational DNA repair [e.g., mei-2 and mei-3 (1838)]. These typically show hypersensitivity to genotoxic agents as well as alterations in mutation and/or mitotic recombination (Table 3). They may be isolated by sensitivity to UV or other mutagens, and they are most commonly called uvs or mus. Mutagen-sensitive mutants with meiotic effects include uvs-3, -5, and 6; mus-7, -8, -9, and 11, and, with the exception of mus-18, all mutants from mus-15 to mus-25.
mei-1 : meiotic-1
IVR. Linked to arg-2 (<1%), probably to the right (1933)
Meiosis is impaired in homozygous crosses. There is no defect in growth or DNA repair. Meiotic divisions occur and many ascospores are produced, but 70-90% are inviable and white. The viable ascospores are usually disomic for one or more linkage groups, indicating high nondisjunction at the first division (1933). Chromosome pairing is defective: axial elements of a synaptonemal complex are present, but a complete complex is rarely seen. Defects at anaphase I and II lead to four-poled second- and third-division spindles (1232). The mei-1 mutation is present in wild-collected strain Abbott 4A (1933), which is an ancestor of many Beadle and Tatum mutants (136). Two mutants obtained in ref. (518), asc(DL95) and asc(DL243), complemented mei-1 and each other and showed no recombination in intercrosses, suggesting allelic complementation and/or a gene cluster. Abnormal disjunction in these crosses resulted from abnormal pairing in meiosis I (519). asc(DL95) resembles mei-1 and may well be allelic; but defects are less extreme. Mutant DL243 differs from mei-1 with a primary defect early, during karyogamy, and pleiotropic effects later, causing high nondisjunction during the second division and postmeiotic divisions (519).
mei-2 : meiotic-2
VR. Between inv (18%) and his-6 (12%) (956, 1933). Linked to mus-11 (5%; 10%) (1840).
Uvs-6 epistasis group (958) (Table 3). Early dominant effects were eliminated by backcrossing to produce the commonly used strain mei-2 ALS181 (1840). A second allele is mei-2 SA60 (956). In homozygous mei-2 crosses, meiotic divisions occur and asci appear to be normal, but they contain few mature ascospores. Chromosome pairing is greatly reduced or absent, resulting in aberrant segregations at anaphase I and often at subsequent divisions [B. C. Lu, cited in refs. (1841) and (1933)]. Meiotic crossing over similarly is reduced and nondisjunction increased, producing aneuploid ascospores, monosomic white inviable spores, and black ones disomic for one or more linkage groups (1841, 1933). Heterozygous crosses produce few white spores, and mitotic recombination is normal (1841). RIP and presumably DNA pairing are not inhibited by mei-2 (663). Crosses homozygous for mei-2 are used in the “sheltered RIP” procedure for determining whether the function of a cloned gene is essential for survival (839, 1335). mei-2 is DNA-repair-defective and sensitive to MMS, histidine, and g rays (1840). In contrast to mei-3, mei-2 has normal spontaneous and UV-induced mutation (958). The mutant affects expression of a peptide that is inducible by DNA damage (911, 913). Epistasis tests with uvs-3 suggest that mei-2 may function in more than one type of repair (958), as is known for some DNA replication genes [e.g., CDC9 (ligase) in yeast], or possibly there may be an overlap between repair types of the Uvs-6 and Uvs-3 epistasis groups.
mei-3 : meiotic-3
IL. Between arg-1 (3%) and T(39311)R. Probably right of eth-1 (1%) and arg-3 (1%) (1469, 1580).
Cloned and sequenced: EMBL/GenBank D29638, L02428, PIR S70629, GenBank NEUMEI3, NEUMEI3A; Orbach-Sachs clones X5D09, X10C02, X10D03, X11H02, X14G03, G1A07, G3B05, G10H05, G11B01.
Uvs-6 epistasis group (958) (Table 3). A homolog of Escherichia coli recA, Saccharomyces RAD51, and Aspergillus uvsC (373, 853, 2141). MEI-3 protein is located specifically in perithecia, but expression also is increased in growing mycelia after UV or MMS treatment (853). The seven known mei-3 alleles are all recessive. The first (N289) was found because it increased the instability of duplications (1469). Some were isolated by MMS sensitivity [e.g., SA10 (958) and SC25, SC29 (520, 521)]. mei-3 (N289) typically is sensitive to ionizing radiation, MMS, histidine, and (slightly) UV (best scored at 39ºC) (1469, 1473). It is also sensitive to hydroxyurea (1840) and mitomycin-C (381). Spontaneous mutation is much increased (mutator phenotype), but further, UV-induced increases in mutation resemble those of wild type (958). Sometimes gives stop-start growth on minimal medium, made more pronounced by histidine (1467). Homozygous barren (1469). Meiosis is blocked in zygotene (1685). Duplication instability is increased (1469), possibly by mitotic deletion. mei -3 may be deficient for mitotic recombination, as is the yeast homolog rad51. Homology to a gene of the yeast Rad52 group suggests that mei-3 and mei-2 may function in recombination and possibly in recombinational repair of double-strand breaks. The mutation affects the expression of a DNA-damage-inducible polypeptide (911, 913).
mei-4 : meiotic-4
IIIR. Linked to leu-1 (12%), probably to the left (1469).
A meiotic-specific mutant without defects in DNA repair (1469, 1840). Isolated from a double-mutant strain with mei-3 (1473). Homozygous barren. Recessive. Expression is highly variable, depending on genetic background (1469). The more extreme genotypes block sexual development at crozier formation or karyogamy (1685). Less extreme genotypes complete a normal first meiotic division but become irregular at later divisions, producing abnormal spores [B. C. Lu and D. R. Galeazzi, cited in ref. 1685)].
mek-1 : MAPK/ERK kinase
Unmapped.
Cloned: Orbach-Sachs clone G09C10 (1267).
Cloned by PCR with degenerate primers encoding conserved domains of MAPK (mitogen-activated protein kinase) and MEKK (MAP/ERK kinase kinase)-conserved domains (1267). The predicted protein shows similarity to products of Saccharomyces MKK1 and MKK2, which are involved in a protein kinase C-responsive MAPK pathway. Proteins of this type phosphorylate MEK proteins in other organisms (1267).
mel-1 : melon-1
VIIL. Left of thi-3 (27%) (1604).
A hemispherical colony is formed similar to bal (1409). Growth is stimulated rather than depressed by sorbose (1407). Cell-wall analysis (507). Photographs (507, 1409). Called col(C-L2b). Not tested for allelism with do.
mel-2 : melon-2
Allelic with bal (1585).
mel-3 : melon-3
III. Linked to leu-1 (17%) (1409).
Grows as a hemispherical colony similar to bal. A modifier gene also is located in III (1409).
mep : methylpurine resistant
Mutants resistant to inhibition by 6-methylpurine (6MP) are potentially useful as markers. Mutants called mep(3) and mep(10) were described by refs. (1542) and (1545) as differing phenotypically. Both have been mapped in IL. Another 6MP-resistant mutation was obtained independently in a T(OY321) strain (1322), mapped in IL between leu-3 (10%) and T(OY321) (“tightly linked”), and shown to be recessive in partial diploids (261). Allelism of the three mutants has not been tested. Because a stock of mep(3) received by R. L. Metzenberg (1322) proved to be heterokaryotic for a nonresistant component, doubt is cast on the validity of the phenotypic diagnosis given next for mep(3). Possibly all three mep mutations may be at the same IL locus.
mep(3) : methylpurine resistant(3)
IL. Left of mat (1322).
Resistant to inhibition by 6MP. The activity of adenine phosphoribosyltransferase (PRTase) (EC 2.4.2.7) is nearly normal in vitro. Adenine uptake is normal. Has low hypoxanthine PRTase, as do mep-2 and ad-8. Selected on 1 mM 6MP sorbose medium (1542). 6MP prevents purple pigment production by ad-3 on low adenine, but 6MP does not prevent pigment production by ad-3A mep(3) double mutants, suggesting that mep(3) is altered in the regulation of de novo purine synthesis. The phenotype is consistent with lowered affinity of glutamine amidotransferase for 6MP as a feedback inhibitor (1545). Not tested for allelism with mep-2 or with adenine mutants. Called Mepr-3 (1542), mep-1 (107). If these results were obtained using the heterokaryotic stock received by R. L. Metzenberg (1322), the mep(3) and mep(10) mutations could be at the same locus.
mep(10) : methylpurine resistant(10)
IL. Linked to leu-3 (0/120) (1546).
Resistant to inhibition by 6MP. PRTase activity is negligible in vitro. Adenine uptake is normal. Purine synthesis is not inhibited by 6MP (1545). Resistance may result from the inability to convert the analog to the nucleotide form. Has low hypoxanthine PRTase, as do mep(3) and ad-8. Selected on 1 mM 6MP sorbose medium. Not tested for allelism with mep(3) or with another IL-linked mep mutation mapped in ref. (261). Called Mepr-10. Called mep-2 in ref. (107).
met : methionine
Auxotrophs designated met are able to use methionine but not cysteine. Some can use the methionine precursors homocysteine and cystathionine (Fig. 45). Mutants able to use cysteine as well as methionine are designated cys. Reviewed in ref. (653). For regulation, see ref. (1874) and entries for individual loci. Methionine starvation of mutants in the met pathway results in decreased DNA methylation (1732).
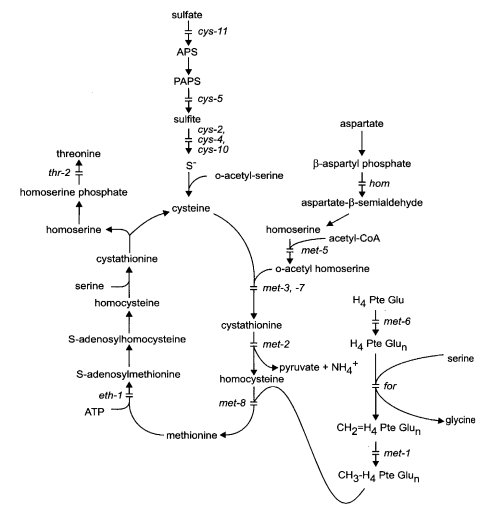
FIGURE 45 The sulfur assimilatory pathway leading from inorganic sulfate to cysteine, methionine, homoserine, and threonine. Mutants that affect specific steps are identified. For the conversion of threonine to isoleucine, see Fig. 39. It is not clear whether the polyglutamylation step controlled by met-6 occurs only at the stage shown. Abbreviations: APS, adenosine 5'-phosphosulfate; H4PteGlu, tetrahydrofolate; PAPS, 3'-phosphoadenosine=5'-phosphosulfate. Adapted from refs. (1284 ) and (1596 ).
met-1 : methionine-1
IVR. Between oxD (3%), T(B362i)L and T(B362i)R, mus-26 (2%), col-4 (4%) (111, 322, 1411, 1490, 1578).
Uses methionine but not homocysteine (893) (see Fig. 45). Lacks methylene tetrahydrofolate reductase (EC 1.5.1.5), and, thus, lacks the coenzyme needed for transmethylating homocysteine (260, 1872, 1873). A report that met-1 also lacks cystathionine synthase (1045) proved to be incorrect, due to the fact that methyltetrahydrofolate is an essential activator for cystathionine synthase (1874). Methylene THF reductase is feedback-inhibited by S-adenosylmethionine (260). Used in heteroallelic duplications from T(S1229) to assay mitotic recombination (113). Methionine starvation decreases DNA methylation (1732).
met-2 : methionine-2
IVR. Linked to ilv-3 (0/129) (1124). Between trp-4 (6%) and pan-1 (4%) (1410).
Uses methionine or homocysteine; accumulates cystathionine (893). Lacks cystathionase II (EC 4.4.1.8) (655) (Fig. 45). Fine-structure map (1413, 1417). Used in major studies of intralocus recombination and its polarity (1413, 1417). Complementation map (1410). Methionine starvation decreases DNA methylation (1732).
met-3 : methionine-3
VR. Between pab-1 (1%), trp-5 (4%) and pk (1%) (13, 212, 569, 673, 2014)
Uses methionine, homocysteine, or cystathionine (893). Putative structural gene for cystathionine-b-synthase (EC 4.2.1.22) (1045) (Fig. 45). This enzyme is also lacking in met-7 (1045). The enzyme is activated by methyl tetrahydrofolate and inhibited by S-adenosylmethionine (1045, 1874). A temperature-sensitive met-3 allele is deficient in DNA methylation (1732).
met-4 : methionine-4
Changed to cys-10 (1414).
met-5 : methionine-5
IVR. Between his-4 (4%) and nit-3 (15%) (212, 1582, 1585).
Cloned: pSV50 clone 18:5B.
Uses cystathionine, homocysteine, or methionine (656, 1411). Defective homoserine transacetylase (EC 2.3.1.31) (1045, 1426) (Fig. 45). Methionine starvation decreases DNA methylation (1732).
met-6 : methionine-6
IR. Between thi-1 (7%; 14%), T(NM103), T(ALS182), T(OY343 and tre (7%), ad-9 (2%; 16%) (890, 1415, 1548, 1580, 1582, 1818, 2123).
Cloned and sequenced: EMBL/GenBank AF005040, EMBL NCAF5040, pSV50 clones 4:11C, 7:10D, 8:1F, 8:4H (68).
Structural gene for folyl polyglutamate synthetase (EC 6.3.2.17). Requires methionine and does not use precursors [(1411; N. Horowitz, cited in ref. (2294)] (Fig. 45). Used to study the polarity of intralocus recombination and to show that polarity with respect to flanking markers is not reversed when the met-6 region is inverted relative to the centromere (1416, 1417). Used to show that methionine starvation decreases DNA methylation (1732). Strains carrying allele 65108, called mac, were reported to differ from other met-6 mutants in having an accessory requirement for adenine and possibly cystine (556), whereas met-6 (35809) and (S2706) are stimulated by adenine only in a CO2-enriched atmosphere [G. Roberts, cited in ref. (1417)]. met-6 (65108) (mac) and met-6 (35809) evidently lack different folyl polyglutamyl synthetase activities (EC 6.3.2.17) (410, 1728). This may be due to alterations in different domains of the same gene product. The cloned met-6+ gene increases the level of folyl hexaglutamates when transformed into met-6 or mac mutants (67). A highly conserved Ser residue has been changed to Pro in allele 35809. Sequence polymorphisms among wild-type strains (68).
met-7 : methionine-7
VIIR. Between qa-2 (<1%), ars (<1%), Cen-VII (<1%) and met-9 (10-4), wc-1 (1%; 4%) (295, 304, 340, 1411, 1418).
Cloned and sequenced: Swissprot MET7_NEUCR, EMBL/GenBank M64066, PIR JQ1524, EMBL NCMET7A, GenBank NEUMET7A; Orbach-Sachs clones G7G03, G17H07.
Uses cystathionine, homocysteine, or methionine [(1411); N. Horowitz, cited in ref. (2294)]. Structural gene for cystathionine g-synthase (EC 4.2.99.9) (427, 1045) (Fig 45). Note that cystathionine-b-synthase (EC 4.2.1.22) is lacking in met-3 (1045). See met-3 for regulation. Apparently contiguous with met-9 by co-conversion. Flanking markers are recombined in most met-7+ met-9+ recombinants (1418). Functionally distinct from met-9, which has active cystathionine synthase (1045) but cannot use homocysteine. No mutants lacking both functions have been isolated. Allele NM251 is suppressible by ssu-1 (1418). Allele K79 is inseparable from reciprocal translocation T(K79) (1580).
met-8 : methionine-8
IIIR. Between ace-2, ff-5 (1%, 4%) and ad-4 (4%), leu-1 (426, 1411, 1546, 1591, 2044).
Cloned and sequenced: GenBank A1391967; EST NC1D10.
Uses methionine but not its precursors (1411). Lacks methyl THF homocysteine transmethylase (EC 2.1.1.13) (260, 1873) (Fig. 45). Methionine starvation decreases DNA methylation (1732).
met-9 : methionine-9
VIIR. Between met-7 (10-4) and wc-1 (1% or 2%) (1418, 1591).
Requires methionine; cannot use precursors (556, 1411, 1418). Co-converted with met-7 (1418). Functionally distinct from met-7. The met-9 mutant retains the met-7+ function, producing cystathionine synthase (1045). Allele NM43 is heat-sensitive (1418). Starvation for methionine decreases DNA methylation (1732).
met-10 : methionine-10
IR. Between his-2, nuc-1, T(AR173)R and lys-4, his-3, ad-3A (1022, 1322, 1469, 1578).
Cloned and sequenced: PIR JC4255, EMBL/GenBank L40806, EMBL NCORF, Genbank NEUORF.
Requires methionine. Encodes a 475-residue polypeptide of unknown function (377). The only known allele is heat-sensitive, with requirement at 34ºC but not at 25ºC (1592). Does not grow at 39ºC even with methionine (1469). Methionine starvation decreases DNA methylation (1732).
meth : methionine
Symbol changed to met.
Methionine Overproduction
See eth-1 (1028).
mfA-1 : mating factor A-1
Allelic with ccg-4.
mfa-1 : mating factor a-1
VR. Linked to leu-5 (0T/18 asci) (1447).
Cloned and sequenced: Orbach-Sachs clones X13E01, X22F11.
Encodes a pheromone precursor expressed in mat a. The sequence suggests an isoprenylated fungal pheromone similar to the a-factor of Saccharomyces (573). For other genes expressed differentially in the two mating types, see ccg-4, eat-1, eat-2, and scp.
mgk-1 : glycogen synthase kinase-1
III. By correlation with cosmids assigned to linkage groups (573).
Cloned: Orbach-Sachs clone G03D09 (1267).
Cloned by PCR with degenerate primers encoding MAPK (mitogen-activated protein kinase)-conserved domains (1267). The predicted protein shows similarity to glycogen synthase kinases, which are conserved in fungi and animals where they have roles in development and gene regulation (1267).
Microconidiation ;
Microconidia, which are uninucleate, are developmentally and morphologically distinct from macroconidia. Microconidia in wild type are obscured by the presence of macroconidia. A simple method has been devised for obtaining large numbers of almost pure microconidia from wild type (1528). Microconidia can also be obtained in large numbers using liquid shake cultures inoculated with macroconidia of the mcm mutant (1251). Morphologically mutant strains pe and dn produce exceptionally high numbers of microconidia, together with macroconidia. Macroconidiation is eliminated in the fl (fluffy) mutant, but microconidiation is not. Double-mutant strains of constitution pe fl and fl; dn have long been used as sources of pure microconidia. For a photograph of microconidiophores, see Fig. 46 (1528). For an SEM photograph of microconidia erupting from hyphae, see ref. (1959). For a TEM photograph of microconidia from pe; fl, see ref. (1230). For a review of Neurospora microconidia, see ref. (1252).
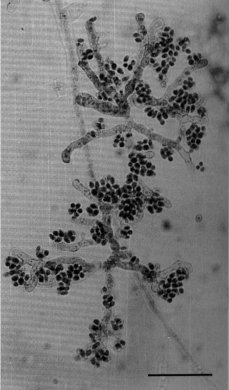
FIGURE 46 Aerial microconidiophores of wild-type N. crassa, formed after the removal of cellophane from the surface of an agar culture following the procedure of Pandit and Maheshwari (1528 ). Stained with acid fuchsin. Groups of microconidia are seen arising from very short protuberances from branches of two microconidiophores. Bar length = 50 mm. From ref. (1528 ).
mig : migration of trehalase
Allelic with tre (2210).
mik-1 : MEK kinase-1
Unmapped.
Cloned: Orbach-Sachs clone G01H02 (1267).
Cloned by PCR with degenerate primers encoding MEKK-conserved domains (1267). The predicted protein shows similarity to the product of Saccharomyces SLK1 (also known as BCK1), which is involved in a protein kinase C-responsive MAPK pathway. Proteins of this type phosphorylate MEK proteins in other organisms (1267).
mik-2 : MEK kinase-2
Allelic with nrc-1.
mip-1 : MIP1-like DNA polymerase
IIIR. Between pro-1 and ad-2. Linked to con-7, trp-1 (1T/18 asci) (1078, 1079).
Cloned and sequenced: EMBL/Genbank AF111068; Orbach-Sachs clone X25C10.
Specifies mitochondrial DNA polymerase g (large catalytic subunit) (EC 2.7.7.7), resembling MIP1 polymerase of Saccharomyces and POL-G of Escherichia coli (1078, 1079).
mlh-1 : mutL homolog-1
IV. Linked to mtr (0/18) (956, 2254).
Cloned and sequenced: Orbach-Sachs clones X14D11, X19B3, X21B5.
Homolog of the gene that encodes the Escherichia coli mismatch repair protein MUTL (2254). RIP-inactivated mutants show a mutator phenotype, are not sensitive to mutagens (UV, MMS, NG, cisplatin II), and are homozygous-fertile (2254).
mo : morphological
A miscellaneous group of mutants differing visibly from the wild type in vegetative morphology. The symbol morph has also been used. Other categories of morphological mutants have been designated col, spco, smco, or moe or have been assigned descriptive names such as bal, fr, ro, sc. For reviews covering morphological mutants and morphogenesis, see refs. (235), (384), (1281), (1353), (1848), (1851), and (2117). Reference (719) describes and gives photographs of numerous morphological mutants. Growth rates and hyphal diameters of 18 morphological mutants are given in ref. (384). For scanning electron microscope photographs of numerous morphological mutants, see refs. (1955), and (1958). A selective enrichment technique for obtaining morphological mutants has been described on the basis of their inability to use acetate and glutamate as sources of carbon and nitrogen (530). Mutations in ropy genes, which affect dynein or related molecular motors, can be obtained by selecting suppressors of cot-1 (247, 1634). crisp mutations can be obtained by selecting suppressors of mcb (247). Mutations in osmotic and spco genes can be obtained by selecting for resistance to dicarboximide fungicides (788-790). The Fungal Genetics Stock Center lists ~30 morphological mutants that have been mapped to a linkage group, but have not been tested for allelism with already mapped morphologicals (700). These are not listed here.
mo-1 : morphological-1
I. Linked to mat (9%) (719).
Growth from ascospores is slow (719).
mo-2 : morphological-2
VII. Linked to for (16%), nt (29%) (719, 1582).
The mycelium is slow-growing and poorly pigmented. Conidia are morphologically abnormal. Recovery from ascospores is poor (1582). Scanning EM photograph (1955).
mo-3 : morphological-3
Allelic with sk.
mo-4 : morphological-4
IIIR. Right of leu-1 (8%). Linked to pro-1 (10%) (719).
Conidiation occurs throughout a slant. Complements col-l4, col-l6, spg (719).
mo-5 : morphological-5
I. Linked to mat (20%) (719).
Few conidia. May make exudate on slant (719).
mo(KH160) : morphological(KH160)
Changed to shg.
mo(P1163) : morphological(P1163)
Changed to dr.
mo(P2402t) : morphological(P2402t)
Changed to un-20.
mod-5 : modifier of permeability-5
VI. Linked to Cen-VI (3% second-division segregation), trp-2 (1973). Probably an allele of cpc-1 in VIL, but not tested for allelism.
Improves the growth on complex media of trp-1, -2, -3, and -4, aro-1, tyr-1, and -3, pt, met-7, and pyr-1; increases sensitivity to 4-methyltryptophan and p-fluorophenylalanine. Recessive in heterokaryons. Attributed to a permeability change that facilitates the entry of metabolites (109, 1973). Scorable on slants of minimal medium + 4-methyl-DL-tryptophan (0.9 mg/ml, autoclaved in medium; tests read at 7 days, 34ºC) (1582). mod-5 differs from cpc-1 (mts) in enabling excellent growth of pyr-1 (H263) on complex media (323).
mod(os-5) : modifier of os-5
Unmapped.
Restores normal morphology and osmotic sensitivity to os-5 while retaining resistance to dicarboximide fungicides (786).
mod(pr) : modifier of partial resistance to killing
IIIR. Between leu-1 and his-7. Outside the recombination block that is associated with Sk-2 (2121).
Enhances the resistance to killing by Spore killer-2 that is conferred by pr(Sk-2) (2121).
mod(sc) : modifier of scumbo
IV. Linked to pan-1 (17%) (930).
Restricts the growth of sc but not of cr-1, fr, bis, sp, or wild type (930).
moe-1 : morphological, environment sensitive-1
Probably an sk allele (1582).
moe-2 : morphological, environment sensitive-2
VI. Linked to trp-2 (14%), probably to the left (719).
Grows with concentric zones on minimal medium and as a restricted colony on glycerol complete medium (34ºC). For photographs of allele R2532, see Figs. 23 and 24 of ref. (719). scot was probably present in the strain of origin.
moe-3 : morphological, environment sensitive-3
IV. Left of pan-1 (17%; 25%), bd (20%) (1821).
Blocks conidial germination at high temperature. Mycelial growth is colonial at high temperature if on dialysis tubing on an agar surface, but is fairly normal if submerged. Shows a strong circadian conidiation rhythm at low temperature (1821). The effect on conidial germination (but not on vegetative growth) is counteracted by high conidial concentration or by CO2 (360, 1821). Histidine is stimulatory, but there is disagreement as to whether it affects germination or vegetative growth (359, 1821). Partially curable by siderophores (ferricrocin). Conidia rapidly lose siderophores on contact with aqueous medium, even at permissive temperatures, suggesting an altered plasma membrane attachment site (360). Called JS134-9.
mom : mitochondrial outer membrane
Changed to tom: translocase outer mitochondrial membrane.
mom38 : mitochondrial outer membrane 38
Changed to tom40.
morph : morphological
Symbol changed to mo.
mpp : mitochondrial processing peptidase
Unmapped.
Cloned and sequenced: Swissprot MPP1_NEUCR, EMBL/Genbank J05484, PIR A36442, EMBL NCMPPX, GenBank NEUMPPX.
Encodes the 57-kDa subunit of mitochondrial processing peptidase (EC 3.4.24.64) (1823). MPP cooperates with PEP (processing enhancing protein, encoded by pep) (860) in the proteolytic cleavage of matrix-targeting sequences from nuclear-encoded mitochondrial precursor proteins. (The 52-kDa subunit is encoded by pep.)
mrp(3) : mitochondrial ribosomal protein (3)
Unmapped.
Cloned and sequenced: Swissprot ODP2_NEUCR, EMBL/Genbank J04432, PIR A30775, GenBank NEURPNUC.
Encodes a mitochondrial protein found both on ribosomes and in membrane fractions. Includes a domain similar to one found in dihydrolipoamide acetyltransferases (1097, 1750). Called mrp-3. Genes encoding at least two other proteins, MRP13 and MRP16, are said to be under study, and an account of a gene called mrp-15 is said to be in preparation (2193).
msh-1 : mutS homolog-1
IL. Linked to cyt-21 (0/18) (956, 2254).
Cloned and partially sequenced.
Homolog of genes that encode Escherichia coli mismatch repair protein MUTS and Saccharomyces MSH1, MSH3, and MSH6 (956, 2254). Called msh-X.
msh-2 : mutS homolog-2
VIIR. Linked to for, frq (0T/18 asci). Right of Cen-VII (1T/18 asci) (1447); (0/18) (956).
Cloned and sequenced: EMBL/GenBank AF030634; Orbach-Sachs cosmids X95H, X179E (956), X20E3, G1H8; YACs 8:E:6, 17:C:11 (162).
Homolog of genes that encode mismatch repair proteins Escherichia coli MUTS and Saccharomyces MSH2 (935, 2254). RIP-inactivated mutants show a mutator phenotype, are not sensitive to mutagens (UV, MMS, NG, cisplatin II), and are homozygous-fertile (2254).
msk-1 : STE20-like MAP kinase-1
I. By correlation with cosmids assigned to linkage groups (573).
Cloned: Orbach-Sachs clone G03H05 (1267).
Cloned by PCR with degenerate primers encoding MEKK-conserved domains (1267). The predicted protein shows similarity to the products of Saccharomyces STE20, SPS1, and CLA4 and S. pombe shk1 and to vertebrate PAK proteins. In other systems, this class of kinases phosphorylates/activates the MEKK proteins to initiate the MAPK/kinase cascade (1267).
mt : mating type
This symbol, long used for mating type, was changed to mat in 1997 (745).
mtr : methyltryptophan resistant
IVR. Between pdx-1 (2%) and col-4 (1%) (196, 1990).
Cloned and sequenced: Swissprot MTR_NEUCR, EMBL/GenBank L34605, S81767, PIR A54551, EMBL NCMTR, GenBank NEUMTR; pSV50 clones 5:10A, 5:3B, 5:4H.
Structural gene for the transport of neutral aliphatic and aromatic amino acids via amino acid transport system I as defined by ref. (513), (1177), (1521), (1990), and (2233) (Fig. 47). Resistant to 4-methyltryptophan (MT) and p-fluorophenylalanine (FPA). Resistance is recessive in heterokaryons and in duplications from T(S1229). Mutation causes alteration in surface glycoproteins (2016). Used extensively for studying transport [(512); reviewed in (2231) and (2233)], mutation (541, 1981, 1982, 2192), intergenic deletion mapping (1688, 2192), and recombination (1692). Forward mutations may be obtained by selection for resistance or for defects in uptake ability; revertants are selected by the ability of an auxotroph to grow on appropriately supplemented medium (1981). Used in one component of a heterokaryon test system for measuring the rate of recessive lethal mutation throughout the genome (1979, 1980) (Fig. 22). Suppressors have been used for selecting other resistance mutants (227, 228, 1067, 1991). FPA resistance is suppressed by am (1335). A light-regulatable al-3::mtr fusion gene, which is FPA-sensitive in the light and FPA-resistant in the dark, was used to select light-regulation mutants (286). Ascospores carrying mtr are slow to darken and mature; up to 50% of young ascospores from heterozygous crosses are white (318, 1582). With probable allele MN18, ascospore viability is improved by adding peptone to the crossing medium when the male parent is added (318). For scoring, medium containing 15 mg/ml FPA is recommended as an alternative to MT (541). Unlike MT, FPA is heat-stable and can be added before autoclaving. Mutants called pmn (=Pm-N, pm n), selected by resistance to FPA, have been shown allelic with mtr [(1776); see also ref. (513)]. Strains originally designated neua, neur, neut (1243) may be mtr alleles. Called mt (1177).
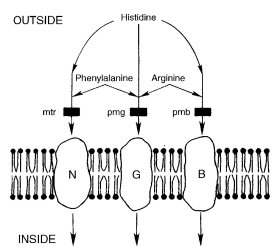
FIGURE 47 A schematic diagram of the three major systems for transport of amino acids across the Neurospora plasma membrane: neutral (N), general (G), and basic (B). Histidine is transported by all three systems. Phenylalanine is transported by the N and G systems, whereas arginine is transported by the B and G systems. Mutations that reduce or eliminate these transport activities are represented by heavy black lines intersecting the arrows and are labeled with the standard Neurospora designations mtr, pmg, and pmb. Whether the systems consist of more than one gene product is not known. N = system I (1521 ), G = system II, and B = system III. From ref. (2017 ), with permission from the National Research Council of Canada.
mts : methyltryptophan sensitive
Allelic with cpc-1 (327, 1081).
multicent
This name has been given to strains that have centromere regions suitably marked for assigning genes or chromosome rearrangements to linkage groups. Strains, markers, and methods are described in ref. (1571). Strains multicent-1, -3, -4, and -5 have been especially useful for mapping chromosome rearrangements, especially translocations. Scoring of marker phenotypes is somewhat more laborious with multicent strains than with alcoy; csp-2, which is therefore preferred for locating point mutants. For ease of crossing and stock-keeping, phenotypically wild type heterokaryons have been constructed of multicent-3, -4, and -5 in combination with the inactive-mating-type helper strain am1 ad-3B cyh-1.
multicent-1
Linkage tester strains (of both mating types) with mat, bal, acr-2, pdx-1, at, ylo-l, and wc-1 marking the centromere-proximal regions of linkage groups I-VII (1556, 1571).
multicent-2
A strain with mat-a un-2, arg-5, thi-4, pyr-1, lys-1 inl, and nic-3 ars-1 marking six linkage groups in OR-like genetic background. This was crossed with the highly polymorphic wild-collected strain Mauriceville-1c A to obtain a kit of progeny for use in RFLP mapping (1334, 1339, 1340) (Appendix 3).
multicent-3
Strains of constitution In(IL;IR)OY323, arg-5, acr-2, pdx-1, at, ylo-1, and wc-1 (1571). These increase efficiency over multicent-1 by introducing the inversion as a crossover suppressor in I and by substituting arg-5 for bal in II. Because the inversion is heterozygous in test crosses, markers in most of linkage group I will show close linkage to mating type.
multicent-4
mat, arg-5, acr-2, psi, at, ylo-1, wc-1 (1571). The temperature-conditional marker psi has been substituted for pdx-1 in IV.
multicent-5
In(IL;IR)OY323, arg-5, acr-2, psi, at, ylo-1, wc-1 (1571). Like multicent-4 but with the inversion present to suppress crossing over of I markers with mat.
mus : mutagen sensitive
This symbol was adopted in 1980 as a general designation for DNA-repair-defective mutants (1839). Locus numbers begin with mus-7 (1010, 1017) to avoid confusion with the previously named genes upr-1 and uvs-1 through -6. All these mutants are sensitive to genotoxic agents other than UV (946, 1838, Table 3). Discrimination of mus, uvs, and mei-2 or mei-3 from wild type in the progeny of crosses is often readily accomplished by simple spot or stab inoculation of sorbose plates containing MMS or another genotoxic agent (see also uvs). These DNA-repair genes plus phr, which is active in the photoreactivation of UV damage, represent several different repair types. Grouping by epistasis in double-mutant strains corresponds well with phenotypic and functional characteristics (946, 1838). Seven epistasis groups have been identified; Table 3 (946).
mus-7 : mutagen sensitive-7
IIR. Between arg-5 (8%; 12%) and nuc-2 (11%) (948, 1017).
Epistasis group Uvs-6 (Table 3). Extremely sensitive to histidine, highly sensitive to MMS, moderately sensitive to nitrosoguanidine and bleomycin, but insensitive to UV, mitomycin C, or ionizing radiation (1010, 1016). Both spontaneous and UV- or X-ray induced mutation are normal (1010). mus-7 resembles mus-16 in showing high MMS-induced mutation and in other common features (954). Homozygous barren (1010), but intercrossed alleles can produce some progeny (0 mus+ /300) (948). Increased mitotic chromosome instability is correlated with sensitivity to hydroxyurea (1835). Function is uncertain, considering the absence of radiation sensitivity. Three similar alleles, (FK116) (1015), (SA1) (948), and (FK107); the latter is used generally.
mus-8 : mutagen sensitive-8
IVR. Linked to mtr (1%). Right of pdx-1 (6%) (1010, 1017).
Cloned and sequenced: Swissprot UBC2_NEUCR, EMBL/GenBank D78372, PIR S71430, Genbank NEUMUS8.
Epistasis group Uvs-2 (1017; Table 3). A homolog of S. pombe rhp6, Saccharomyces RAD6 (1952), coding for a ubiquitin-conjugating enzyme. When transferred into Saccharomyces, the Neurospora gene shows good complementation with RAD6 for defects in damage-induced mutagenesis, but only partial complementation for sporulation defects. Highly sensitive to UV, mitomycin C, and MMS, moderately sensitive to X rays and nitrosoguanidine, and not sensitive to inhibition by hydroxyurea or histidine (1010, 1836). Good conidial viability, but growth is reduced on minimal medium supplemented with sorbose, possibly reducing the recovery of mutants selected on that medium. Spontaneous and UV-induced mutations are moderately decreased, but X-ray mutagenesis is normal (1010). Homozygous barren (1017). Perithecia and beaks from mus-8 ´ mus-8 are well-developed, but multiple postmeiotic mitoses occur without chromosome replication, resulting in asci that contain many small nuclei, each with only a few chromosomes similar to polymitotic in maize (1671). The products of mus-8 and uvs-2 may interact, as do products of the corresponding yeast genes RAD6 and RAD18 (the homolog of uvs-2) (2099). Allele FK108 is usually analyzed.
mus-9 : mutagen sensitive-9
IR. Between cyh-1 (18%) and al-2 (6%) (1010).
Epistasis group Uvs-3 (Table 3). Highly sensitive to UV, MMS, MNNG, and histidine, moderately sensitive to X rays, mitomycin C (1010), and 5-azacytidine (662), and also sensitive to bleomycin and hydrogen peroxide, especially during growth (1016). Spontaneous mutation is high (mutator phenotype); there is little or no increase in mutability by UV or X rays (1010). Homozygous sterile (1017). Mitotic chromosome instability is increased; this is correlated with increased sensitivity to hydroxyurea (1835). Levels of a nuclease that is secreted or that leaks from colonies on sorbose DNA agar are reduced (1014) (see nuh). Resembles uvs-3 in most respects. Like uvs-3, mus-9 is lethal in combination with uvs-6 (1012). Three alleles are known (1015). Allele FK104, which has usually been used, has low conidial survival and shows the most pronounced mutagen sensitivity (1017).
mus-10 : mutagen sensitive-10
VIIR. Right of met-7 (7%) (1017)
Epistasis group Uvs-6 (1012; Table 3). Moderately sensitive to UV, MMS (1010), and 5-azacytidine (662). Slightly sensitive to histidine. Insensitive to X rays, MNNG, and mitomycin C. Spontaneous mutation is normal, as is mutation induced by UV or X-rays (1010). Homozygous fertile, though less fertile than wild type (1017). Chromosome stability is normal (1835). Allele FK110 was reported to show lower radiation survival than allele FK105. The difference was reduced by back-crossing.
mus-11 : mutagen sensitive-11
VR. Between ad-7 (6%; 11%), pab-2 (7%) and inv (4%), un-9 (10%), mei-2 (5%; 10%), his-6 (6%). Linked to refs. (1016), (1017), and (1828).
Cloned and partially sequenced: Orbach-Sachs cosmid X198E (961).
Epistasis group Uvs-3 (1012) (Table 3). Two alleles, FK111 and FK117, are both highly sensitive to MMS, extremely sensitive to histidine (1015), significantly sensitive to X rays, g rays, and bleomycin, and slightly sensitive to mitomycin C and hydrogen peroxide (1010, 1016). Moderately sensitive to MNNG (1835) and 5-azacytidine (662). Similar to uvs-3 except for greater sensitivity to MMS and histidine. UV survival, which is moderately reduced, is also probably similar (1010, 1015). The mutant generally used (FK111) is more difficult to work with, being more variable (possibly a higher mutator effect). Conidial viability is similar for the two alleles. Spontaneous mutation is high, increasing the genetic variation of strains in the course of experiments (1012). (In epistasis tests, greater MMS sensitivity of one double-mutant strain compared to single mutants can be attributed to spontaneous mutation.) There is little or no increase in mutability by UV or X rays (1010). Homozygous crosses are barren. Sterility is dominant in crosses to linkage tester strains, but some progeny have been obtained from crosses of mus-11 to prototrophic linkage tester heterokaryons. Mitotic chromosome instability is increased, correlated with sensitivity to hydroxyurea (1835).
mus-12 : mutagen sensitive-12
VR. Left of inl (10%) (520).
Growing mycelia are highly sensitive to MMS but not to histidine (520). Conidia treated in liquid are sensitive to X rays, but not to MMS or UV (521). Spontaneous mutation is increased >10-fold. One mutant, SC15.
mus-13 : mutagen sensitive-13
IR. Right of al-2 (18%) (520).
Growing mycelia are slightly sensitive when grown on MMS medium. Growth is poor on minimal medium. Conidia are not sensitive to MMS, radiation, or histidine. Fertility is normal (521). One mutant, SC28.
mus-14 : mutagen sensitive-14
VI. Linked to lys-5 (520).
Growing mycelia are moderately sensitive to MMS, but conidia are not. Insensitive to UV, X rays, nitrosoguanidine, and histidine (521, 1836). Poor growth. Homozygous fertile (520). Spontaneous mutation is reduced (521). One mutant, SC3.
mus-15 : mutagen sensitive-15
IL. Between arg-1(4%) and arg-3 (<2%) (948, 1836).
Sensitive to MMS and MNNG, slightly sensitive to UV, and not sensitive to histidine. Homozygous barren (948). One mutant, SA7.
mus-16 : mutagen sensitive-16
VL. Between caf-1 (4%) and lys-1 (9%) (954).
Isolated as a mutant sensitive to nitrogen mustard (86). Defective in DNA-DNA and DNA-protein cross-link repair. Highly sensitive to MMS and MNNG, slightly sensitive to histidine and HU, and not sensitive to UV, γ rays, or mitomycin C (confirming that mitomycin C does not form cross-links in Neurospora DNA; 954). Spontaneous mutation is normal; MMS-induced mutation is increased 10-fold at low doses. Homozygous sterile. Mitotic chromosome instability is increased (954). One mutant, JMB15.1
mus-17 : mutagen sensitive-17
IVR. Between pan-1 (<2%), and cys-4 (25%) (945).
Low sensitivity to MMS, not sensitive to UV or X-rays. Homozygous barren [summarized in ref. (1836)]. Two alleles, SA17 and SA21 (945).
mus-18 : mutagen sensitive-18
VL. Linked to rDNA (0T/17 asci), T(AR190) (£3%), con-2 (2T/17 asci), caf-1 (20%) (959, 1447)
Cloned and sequenced: EMBL/GenBank D11392, GenBank NEUUVE, PIR S55262.
Epistasis group Mus-18. (Table 3). A new type of repair gene, coding for UV endonuclease. Specific for pyrimidine dimers of both types: cyclobutane-pyrimidine dimers (CPDs) and TC(6-4) photoproducts. Cloned by transformation of highly UV-sensitive Escherichia coli cells (Dphr; DuvrA and DrecA) with Neurospora cDNA in a bacterial expression vector, using selection for increased UV resistance (2253). Homolog found in fission yeast (2039). The cloned Neurospora mus-18+ gene complements mus-18 SA8B (2253). Expressed Neurospora cDNA increases UV resistance of yeast RAD1 and RAD2 mutants and human XPA cells (2253). Expressed cDNA in Escherichia coli lacking apurinic/apyrimidinic nucleases increases resistance to MMS and t-butyl hydroperoxide (1023). A RIP-produced disruption of mus-18+ is phenotypically mus-18 (2253). Mutant mus-18 alleles show very low but exclusive UV sensitivity (959, 2253) and considerable deficiency for the removal of both types of UV pyrimidine dimers. Spontaneous mutation is normal, but UV-induced mutation is greatly increased. Fertility is normal. Conidial viability is high. Synergistic interaction for reduced UV survival in double-mutant strains with all UV-sensitive mutations tested. The double mutant of mus-18 with mus-38 (the yeast RAD1 homolog) is extremely UV-sensitive and does not show any UV-dimer excision (855, 960).
mus-19 : mutagen sensitive-19
IR. Between aro-8 (8%) and un-18 (4%) (945).
Sensitive to MMS, MNNG, g rays, and histidine. Normal UV survival. Homozygous sterile. Two alleles, SA9 and SA19 (945, 1836). [In ref. (1836), SA6 was listed incorrectly as a mus-19 allele, where in fact SA9 was intended (945)].
mus-20 : mutagen sensitive-20
IIIR. Between trp-1 (8%) and phe-2 (18%) (948).
Highly sensitive to MMS and MNNG, slightly sensitive to histidine, and not sensitive to UV or g rays (948). Highly sensitive to 5-azacytidine (662). Homozygous sterile, but a few mature perithecia may be produced (948). One allele, SA2.
mus-21 : mutagen sensitive-21
IIIR. Linked to trp-1 (20%) and T(1) ylo-1 in alcoy (17%) (1016). Right of ad-2. Not allelic with mus-20 (945).
Moderate-to-high MMS sensitivity; low-to-moderate sensitivity to histidine and bleomycin, and not sensitive to UV or mitomycin C (1015, 1016). Good conidial viability. Homozygous barren. Allele SC10 is sensitive to X rays, slightly sensitive to UV, and clearly sensitive to histidine at 37ºC but not at 25ºC. Six alleles: FK120, FK121, and FK127 with similar phenotypes (1015); FK131 and FK132 differing slightly (950); and SC10, which shows high spontaneous mutation and is female-sterile (521).
mus-22 : mutagen sensitive-22
IR. Left of cyh-1 (2%) (945).
Sensitive to MMS, but not to UV, mitomycin C, or histidine. Few perithecia are produced in homozygous crosses (1836). One mutant, SA22.
mus-23 : mutagen sensitive-23
IIR. Between fl (<1%; 2%) and trp-3 (9%) (85, 2190).
Cloned and sequenced: GenBank AB002530; pMOcosX clone X24:11A, Orbach-Sachs clone X24A11 (2190).
Epistasis group Uvs-6 (Table 3), other members of which have roles in recombinational repair. Homologous to Saccharomyces MRE11 and S. pombe rad32, which interact with RAD50-like genes in double-strand-break repair and nonhomologous recombination (946, 2190). The original mutant allele, SA23, was isolated as histidine-sensitive by filtration enrichment in histidine growth medium. It is highly sensitive to a variety of mutagens, including UV, MMS, MNNG, t-butyl hydroperoxide, 4-NQO, histidine, and hydroxyurea (2190). X-ray sensitivity tests have not been reported. Mutants of both yeast homologs are X-ray- and MMS-sensitive. However, only S. pombe rad32 resembles mus-23 in also being sensitive to UV, suggesting that mus-23 may have a similar function in nonhomologous recombinational repair of DNA. The mus-23 mutant is defective in meiosis and ascospore formation in homozygous crosses. Expression is induced by UV or MMS (2190), similar to other members of the Uvs-6 group (947). The double mutant mus-23; uvs-3 is inviable, similar to uvs-6; uvs-3 (2190).
mus-24 : mutagen sensitive-24
IIL. Left of pyr-4 (8%) (945).
Sensitive to MMS, 4NQO, mitomycin C, and MNNG. Homozygous barren (1836). Mutant allele SA24.
mus-25 : mutagen sensitive-25
VIIC. Between met-7 (4%; 6%) (1016) and un-10 (828).
Cloned.
Epistasis group Uvs-6 (Table 3). Homologous to yeast RAD54, a gene involved in homologous recombination with sequence conserved from fungi to humans (1483). Two alleles, SA3 and FK123. Both are highly sensitive to histidine, moderately sensitive to MMS, MNNG, and g rays, and not sensitive to UV or mitomycin C. At least FK123 is slightly sensitive to bleomycin (1016). Homozygous barren.
mus-26 : mutagen sensitive-26
IVR. Between met-1 (3%) and col-4 (3%), arg-2 (2%). Linked to mus-8 (5%, probably to the left) (949).
Assigned to epistasis group Upr-1 (949, 957) (Table 3). However, a subgroup of Uvs-2 is not ruled out. Highly sensitive to UV and 4-NQO, but nearly normal for g rays, MMS, MNNG, and mitomycin C, suggesting a defect in some type of UV excision repair. UV survival curves plateau at moderate UV doses, a characteristic of upr-1 (949). Decreased photoreactivation repair after UV damage is evident mainly at UV doses giving <10% survival. Dimer excision is normal, however (949, 957). Spontaneous mutation is close to control levels. Unexpectedly, UV-induced mutation in the standard forward-mutation tests was lower than in wild type (949) rather than being increased, as is typical for highly UV-sensitive mutants such as uvs-2 (494, 497). UV reversion in congenic strains similarly showed reduced induction at high UV dose for mus-26 and upr-1, but increased rates for uvs-2. Homozygous fertile. One mutant mus-26 allele is known, SA3B. This is unrelated to SA3, which is a mus-25 allele.
mus-27 : mutagen sensitive-27
IIR. Right of nuc-2 (16%) (1016).
Highly sensitive to g rays (but less so than uvs-6), MMS, bleomycin (more so than uvs-6), and histidine, sensitive to UV only at very high dose (10% survival of mus+), and insensitive to mitomycin C. Homozygous fertile (1015, 1016). One mutant, FK124.
mus-28 : mutagen sensitive-28
VL. Left of lys-1 (10%) (1016).
Low sensitivity to UV, g rays, MMS, histidine, and bleomycin. Normal viability and fertility (1015, 1016). One mutant, FK118.
mus-29 : mutagen sensitive-29
VIL. Between chol-2 (14%) and lys-5 (15%) (1016).
Moderately sensitive to MMS and histidine, low but consistent sensitivity to g rays and bleomycin, and not sensitive to UV or mitomycin C. Reduced conidial viability. Homozygous sterile (1015, 1016). One mutant, FK119.
mus-30 : mutagen sensitive-30
IVR. Between mtr (8%) and trp-4 (4%) (1016).
Sensitive to MMS, but not to UV or histidine. Homozygous fertile. One mutant, FK115.
mus-31 : mutagen sensitive-31
IR. Between Cen-I and ad-3B (7%) (950).
Sensitive to MMS, but not to UV. Low fertility in homozygous crosses (950). One mutant, SA11.
mus-32 : mutagen sensitive-32
IR. Between mus-31 (3%) and ad-3B (4%) (950).
Sensitive to UV and MMS (950). One mutant, SA32.
mus-33 : mutagen sensitive-33
VIIR. Between met-7 (10%) and un-10 (8%) (950).
Sensitive to UV and MMS (950). One mutant, SA33.
mus-34 : mutagen sensitive-34
VR. Between leu-5, ure-2 and am, his-1 (3%) (950).
Sensitive to UV and MMS (950). One mutant, SA18.
mus-35 : mutagen sensitive-35
VIIL. Right of nic-3 (<9%) (950, 956).
Sensitive to MMS, but not to UV (950). One mutant, SA50.
mus-36 : mutagen sensitive-36
IVR. Between pan-1 and uvs-2 (950).
Sensitive to MMS, but not to UV (950). One mutant, SA51.
mus-37 : mutagen sensitive-37
V. Between lys-1 and cyh-2 (950).
Sensitive to MMS, but not to UV (950). One mutant, SA53.
mus-38 : mutagen sensitive-38
IL. Left of un-5 (7%), leu-3 (10%) (960). Linked to nit-2 (0/18) (855, 1447)
Cloned and sequenced: EMBL/GenBank AB009461.
Epistasis group Mus-38 (Table 3). Obtained using degenerate PCR of conserved regions from Saccharomyces RAD1 and RAD1 homologs (855). A mutant allele obtained by RIP was called rad1Nc and was used for mapping (855, 1447). A second allele, SA56, was obtained by UV induction in the highly UV-mutable, but only slightly UV-sensitive, strain mus-18, which is UV-endonuclease-defective. This was identified by increased UV sensitivity using filtration enrichment following UV treatment, photoreactivation repair, and periods of dark repair (liquid holding) (960). The two mutants (rad1Nc and SA56) do not complement in heterokaryons and are similar in sensitivity to UV, 4-NQO, mitomycin C, and cisplatin (855). Both show inefficient survival recovery with photoreactivation after UV. Spontaneous mutation is at the wild-type level. UV-induced mutation is increased 6-fold, as is typical for nucleotide excision repair mutants. mus-38 is in the same epistasis group with mus-40, the Neurospora homolog of yeast RAD2, and presumably they function similarly in nucleotide excision repair. mus-38 interacts synergistically with all previously described UV-sensitive mutants. Double mutants with mus-18 (which is UV-endonuclease-deficient and UV-excision-defective) are supersensitive to UV and completely defective in the release from UV-irradiated DNA of thymine dimers of both types: cyclobutane-pyrimidine dimers (CPDs) and TC(6-4) photoproducts (855) (see also mus-40). These findings explain the puzzling inconsistency that whereas mus-18 encodes for a very active UV endonuclease, mus-18 mutants are barely sensitive to UV. Because two alternative excision mechanisms exist, mutants of one or the other excision repair type could be compensated for by efficient repair provided by the other [as is also found in bacteria for Micrococcus luteus, and in fungi for Schizosaccharomyces pombe (2280)].
mus-39 : mutagen sensitive-39
VI. Right of ylo-1 (3%; 8%) (950). Linked to mus-29 (10%).
Sensitive to MMS and histidine (950, 1015). One mutant, FK133.
mus-40 : mutagen sensitive-40
IL. Linked to pzl-1 (0/18). Between cyt-21 (1/18) and fr, nit-2 (3/16) (945, 1447).
Cloned (854).
Epistasis group Mus-38. With mus-38, mus-40 establishes a nucleotide excision repair epistasis group in Neurospora; this is the main UV-repair type in many organisms, including Escherichia coli and Saccharomyces cerevisiae. Neurospora mus-38 is a homolog of Saccharomyces RAD1. Obtained using degenerate PCR of nucleotide excision repair gene RAD2 and homologs (854). A mutant obtained by RIP is moderately sensitive to UV but is not sensitive to MMS or X rays. UV excision is defective. Double mutants with all other UV-sensitive mutations show additive or synergistic interactions; those with mus-18 are supersensitive to UV and are completely defective in release from UV-irradiated DNA of thymine dimers of both types: cyclobutane-pyrimidine dimers (CPDs) and TC(6-4) photoproducts (854).
mus(SA5) : mutagen sensitive SA5
Unmapped.
Extremely sensitive to UV and MMS. Sterile in heterozygous crosses, therefore, not mapped by genetic methods (948).
mus(SA6) : mutagen sensitive SA6
IIIR. Left of ad-2 (5%), trp-1 (8%) (948).
Sensitive to MMS, MNNG, g rays, and histidine. Normal UV survival. Reduced conidial viability. Homozygous barren (948). Incorrectly listed in ref. (1836) as a mus-19 allele (945).
mus(FK125) : mutagen sensitive FK125
IV or V. Linked to T(R2355), cot-1 (32%). Not linked to mus, uvs, or many other markers on IV and V (1015).
Homozygous fertile. Conidial viability is very low (2-20%). Moderately sensitive to MMS and histidine, but not to UV (1015).
mus(FK128) : mutagen sensitive FK128
Unlinked to csp-2 or to alcoy markers, unlike all mapped mus and uvs genes (1015).
Sensitive to MMS and histidine, but not to UV, or mitomycin C. Homozygous barren. Good conidial viability. Expression of mutagen sensitivities varies in different ascospore isolates. The original strain and the more sensitive isolates possibly contain two interacting mutations (1015).
mus(SC17) : mutagen sensitive SC17
V. Left of inl (27%) (520).
Sensitive to MMS, but not to histidine. Sensitivity is shown by mycelium but not by conidia, and only after preincubation at 15ºC. Growth is cold-sensitive on minimal medium (520).
mus(SC28) : mutagen sensitive SC28
Changed to mus-13.
mut-1 : mutator-1
IVR. Linked to trp-4 (<3%) (541).
Spontaneous mutation rates of mtr and trp-2 are increased 10- to 80-fold (541). The mutations are predominantly ̵1 frameshifts.
Return to the 2000 Neurospora compendium main page