nada : NAD(P)ase
IV. Left of ad-6 (18%) (1457).
Structural gene for nicotinamide adenine dinucleotide (phosphate) glycohydrolase. Normal morphology. Recessive in heterokaryons. Allele 62ts is temperature-sensitive, with altered substrate affinity (1457). Identified by the plaque test, using Haemophilus influenzae. Used in a study of glutamic acid decarboxylase during conidial germination (382).
nap : neutral and acidic amino acid permeability
VR. Linked to inl (15%) (982). Right of ure-2 (32%) (2230).
Cloned and sequenced: PIR S47892, EMBL/GenBank AF001032.
Encodes an amino acid permease. Selected as resistant to ethionine + p-fluorophenylalanine (982). Causes reduced amino acid uptake by neutral, basic, and general systems. Also causes reduced uptake of uridine and glucose. The defect is not in amino acid-binding glycoproteins (1695). See ref. (2230) for aspartate uptake and resistance to inhibitors. Scored by spotting a conidial suspension on minimal medium containing 1.5% sucrose, agar, 0.3 mM ethionine, and 0.02 mM FPA.
nd : natural death
IR. Between cys-9 (<2%; 3%) and T(4540)R, thi-1 (<2%; 4%) (162).
Progressive deterioration of mycelial growth is followed by abrupt, irreversible cessation (1903). Manifest on race tubes or after sequential transfers of macroconidia to slants. Rapid degradation of the mitochondrial chromosome results from hyperactivation of recombination between direct repeats (174, 1869). Hypersensitive to sorbose. Conidia die rapidly on slants at 4ºC (1397). Recessive. Stocks are maintained as balanced heterokaryons. An aged strain can be rejuvenated through heterokaryosis or by crossing to nd+ (1903). Mutant nd strains free of modifiers initially grow at the wild-type rate (1397). Used to examine hypotheses of senescence based on faulty protein synthesis (1183) and lipid auto-oxidation with free radical reactions (1391). Not to be confused with mitochondrial genes symbolized ND or ndh, which encode subunits of NADH ubiquinone oxidoreductase (see Appendix 4). For another Mendelian mutant that results in senescence, see sen-1.
ndc-1 : nuclear division cycle-1
VR. Left of arg-4 (2%), inl (6%) (1895).
A heat-sensitive conditional mutant that fails to grow at 34ºC. Recessive. The division cycle is blocked just before the initiation of DNA synthesis, when spindle pole bodies are duplicated but not separated. The effect is not nucleus-limited in heterokaryons. Used to show that asynchronous germination of macroconidia results from the arrest of nuclei that are at different stages of the division cycle when conidia became dormant and that few nuclei are in S-phase at any one time during conidiation (1895).
ndh : NADH dehydrogenase
Used as symbol and name for the seven mitochondrial genes that encode subunits of the 700-kDa NADH:ubiquinone complex of the inner mitochondrial membrane, known as respiratory complex I. See ref. (937) and Appendix 4. (These mitochondrial genes have commonly been symbolized ND.) The nuclear genes that encode the remaining subunits of complex I are called nuo: ;NADH:ubiquinone oxidoreductase. The symbol ndh is not used for nuclear genes.
ndk-1 : nucleotide diphosphate kinase
VR. Between al-3 (10%; 22%) and his-6 (22%; 24%) (1485, 1486).
Cloned and sequenced: GenBank D88148.
Encodes a 15-kDa protein homologous to nucleoside diphosphate kinase (EC 2.7.4.6) of other organisms. Shows NDK activity and protein kinase activity. Not regulated at the transcriptional level by WC-1 or WC-2 (1489). Mapping is based on a mutant allele ndk-1P72H, previously called psp or ps15-1, which was discovered as a segregant from wc-1 strain FGSC 3528 (1485).
ndp64 : NAD(P)H dehydrogenase 64
IVR. Linked to Fsr-4 (0T/18 asci), Tel-IVR (1T/18 asci) (1318).
Cloned and sequenced: EMBL/GenBank AJ236906.
Encodes a 64-kDa mitochondrial NADH dehydrogenase (EC 1.6.5.3) associated with the inner mitochondrial membrane, perhaps in the outer face. The product of ndp64 is not a subunit of complex I (1318). Called p-64.
neu : neutral amino acid transport
A synonym of mtr: methyltryptophan resistant, which has precedence.
nic : nicotinic acid
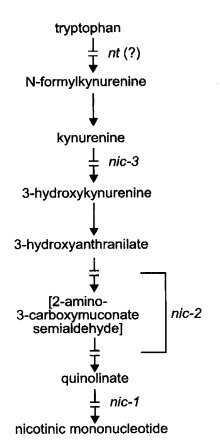
Because of permeability, nic mutants are preferably supplemented with nicotinamide rather than nicotinic acid at most medium pH values (192). To obtain good recovery of some nic mutants from crosses, crossing media should be supplemented with nicotinamide at levels 10-fold higher than those required for growth, even when the protoperithecial parent is nic+ (1548, 1970). nt is best treated as a nic mutant for purposes of growth and scoring. For the biosynthetic pathway, see Fig. 48. For regulation, see refs. (234), (706), (1178), and (1813).
nic-1 : nicotinic acid-1
IR. Between In(OY323)R, lys-3, ace-3 (<1%) and T(UK2-26)R, os-1 (10%; 29%) (9, 115, 278, 1122, 1548, 1578, 1592, 1971).
Cloned (25).
Uses nicotinic acid or nicotinamide, but not precursors (192, 195). Accumulates quinolinic acid (195) (Fig. 48). Affects nicotinic acid synthetase (nicotinate phosphoribotransferase, EC 2.4.2.11). Used to study intralocus recombination (1971). Called the q locus.

FIGURE 48 The pathway from tryptophan to nicotinic mononucleotide, showing sites of gene action (191 , 195 , 702 , 2265 ). The enzymatic reactions between 3-hydroxyanthranilate and nicotinic mononucleotide have not been demonstrated directly in Neurospora. From ref. (1596 ), with permission from the American Society for Microbiology.
nic-2 : nicotinic acid-2
IR. Between ad-3B (4%) and tyr-2, T(Y112M4i)R, ace-7 (4%; 7%) (492, 1122, 1578).
Grows on nicotinic acid, nicotinamide, or high concentrations of quinolinic acid (192, 2265). Cannot use kynurenine, hydroxykynurenine, or hydroxyanthranilic acid (191, 2265). Accumulates 3-hydroxyanthranilic acid (191) (Fig. 48). Affects 3-hydroxyanthranilate 3,4-dioxygenase (EC 1.13.11.6). Aging cultures accumulate red-brown pigment in the medium. Used to study intralocus recombination (1972). Translocations T(4540) and T(S1325) are inseparable from nic-2 (1578, 1972, 1975).
nic-3 : nicotinic acid-3
VIIL. Between do (3%), spco-4 (1%) and lacc (14%), thi-3 (9%; 27%), csp-2 (16%; 22%) (1017, 1582, 1585, 1592).
Uses nicotinic acid, nicotinamide, 3-hydroxyanthranilic acid, 3-hydroxykynurenine, or high concentrations of quinolinic acid (191, 2265). Accumulates a-N-acetylkynurenine; blocked in conversion of kynurenine to 3-hydroxykynurenine by kynurenine-3-monooxygenase (EC 1.14.13.9) (2265) (Fig. 48). Pyridine nucleotide levels (234).
nik-1 : nonidentical kinase-1
An os-1 allele (1844).
nik-2 : nonidentical kinase-2
IIR. Linked to arg-12 (0/17) (1447).
Cloned and partially sequenced: EMBL/GenBank U50263, U50264.
Encodes domains homologous to an SLN1-like histidine kinase (31). The null allele has no observable phenotype (30).
nim-1 : never in mitosis-1
Unmapped.
Cloned and sequenced: Swissprot NIM1_NEUCR, EMBL/GenBank L42573, EMBL NCNIM1A, GenBank NEUNIM1A.
The protein product is homologous to NIMA of Aspergillus nidulans (EC 2.7.1.-), which encodes a cell- cycle, G2-specific protein kinase. Complements Aspergillus nimA functionally (1653).
nit : nitrate utilization
Mutants called nit cannot use nitrate as the nitrogen source, but require ammonia or other sources of reduced nitrogen. Conveniently scored on synthetic crossing medium (2208), in which nitrate is the sole nitrogen source. Also scorable on slants by pH change when grown with ammonium nitrate as the nitrogen source and bromcresol purple (20 mg/ml) as the indicator (1551). In most crosses, nit can be used as the fertilizing parent; in crosses where a nit mutant is required as female parent, crossing medium can be altered by substituting ammonium nitrate for potassium nitrate (320). Nitrite is toxic at low pH; test media containing nitrite should be neutralized, and the nitrite should preferably be filter-sterilized (1947). For a summary of nutritional requirements based on various authors, see ref. (2101). nit-1, nit-7, nit-8, and nit-9 involve a molybdenum-containing cofactor common to nitrate reductase and xanthine dehydrogenase (1155, 2101, 2102) (Figs. 8, 49, and 50). For a review of nitrate assimilation, see ref. (722). For regulation, see reviews in refs. (1282), (1283), (1285), and (1286). See also gln-1, nmr.
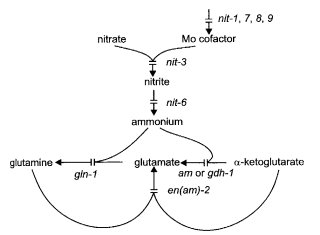
FIGURE 49 The nitrate reduction pathway showing sites of gene action. The am gene
specifies the
NADP-specific glutamate dehydrogenase, and am mutants do not alter the NAD-specific
enzyme, which is specified
by gdh-1
(353 ,
566 ,
639 ,
1027 ,
1155 ,
1781 ,
1948 ,
2102 ).
Modified from ref. (
1596 ),
with permission from the American Society for Microbiology.
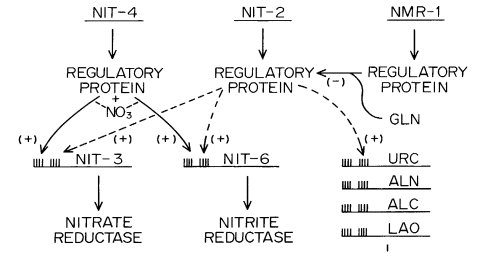
FIGURE 50 The nitrogen regulatory circuit of N. crassa. Expression of genes that encode nitrogen metabolic enzymes only occurs upon nitrogen catabolite derepression and simultaneous induction by a pathway-specific metabolite. The repressing metabolite appears to be glutamine, although the nature of the factor with which it interacts is unknown. NIT-2 is a sequence-specific DNA-binding protein with a single zinc finger that acts globally to activate the expression of many structural genes that encode enzymes of nitrogen metabolism. NMR is a negative-acting regulatory protein that is required to establish nitrogen repression, and it appears to act by binding directly to NIT-2 to inhibit its function. NIT-4 is a pathway-specific positive-acting regulatory factor that has a zinc cluster DNA-binding motif. NIT-4 interacts with NIT-2 to turn on the expression of nit-3 and nit-6. Other specific positive-acting factors are required to express genes encoding enzymes for other nitrogen pathways. URC signifies uricase, for which the structural gene has not yet been identified in Neurospora. uc-5 and ud-1 also are subject to general nitrogen metabolite regulation. Original figure from G. A. Marzluf.
nit-1 : nitrate nonutilizer-1
IR. Between ad-9 (3%; 15%), Tp(T54M94)M and cyh-1 (6%) (890, 929, 1592).
Cannot use nitrate or hypoxanthine as the nitrogen source, but uses nitrite, ammonia, or amino acids (1950). Does not prevent the formation of nitrate reductase apoprotein (1948), but lacks molybdopterin (1095), the molybdenum-containing cofactor common to nitrate reductase and xanthine dehydrogenase (1155, 1441) (Figs. 8 and 49). The nitrate reductase in nit-1 extracts does not catalyze the complete electron transport sequence from NADPH to NO3̵, but it does catalyze the initial part of this sequence if a suitable electron acceptor such as cytochrome c is provided (1948). See ref. (385) for a model of the interaction of nit-1 and nit-3 gene products. For regulation, see Fig. 50 and refs. (442), (1948), and (1950).
nit-2 : nitrate nonutilizer-2
IL. Between un-5 (2%), T(39311)L and lah-1, In(OY323)L, leu-3 (12%, 18%) (115, 1578, 1592, 2041).
Cloned and sequenced: Swissprot NIT2_NEUCR, EMBL/GenBank M33956, PIR A34755, EMBL NCNIT2, GenBank NEUNIT2; pSV50 clone 6:9H, Orbach-Sachs clone G1C05.
Encodes a sequence-specific DNA-binding protein that acts globally to activate many structural genes that are involved in nitrogen metabolism (Fig. 50). The mutant cannot use nitrate, nitrite, purines, or most amino acids as a nitrogen source, but will grow on ammonia, glutamine, or glutamate. Shown to bind GATA sequences (807, 2251). The DNA-binding domain has been characterized (687, 688, 690) as having the DNA sequences to which NIT-2 binds (374-376, 626). The nit-2 mutant is missing (or has severely reduced levels of) nitrate reductase, nitrite reductase, uricase, allantoinase, allantoicase, L-amino acid oxidase, general amino acid permease, extracellular protease, an intracellular neutral PMSF-sensitive protease, and an extracellular RNase (443, 613, 831, 1198, 1712, 1951). NIT-2 does not appear to be necessary for the production of xanthine dehydrogenase (770) or acid phosphatase (806). Levels of glutamate dehydrogenases also are affected (442), as are uracil and uridine uptake (266) and levels of nucleotide pools (1523). Prevents leaky growth of am on minimal medium (320). Allele K31 (called pink) originated in Neurospora sitophila and was introgressed into Neurospora crassa (638); the protein product of K31 may show altered mobility (807). Heterozygosity for the closely linked synaptic sequence reduces recombination in nit-2 (313), which is subject to regulation by rec-1 (324). Function of a nonsense (amber) mutation is restored by ssu-1 (1610). Both nit-2 and nit-4 must be functional for the nitrate utilization genes nit-3 (encoding nitrate reductase) and nit-6 (encoding nitrite reductase) to be activated under inducing conditions (1285), and the direct interaction of NIT-2 and NIT-4 is necessary for the strong activation of gene expression (625). Nuclease-hypersensitive sites in NIT-3 depend on NIT-2 and NIT-4 (229). A cosmid that rescues the defective phenotypes of either nmr or nit-2 has been identified (1949). Transformation of Aspergillus nidulans with the nit-2 gene complements some of the defective phenotypes of areA mutants (450); nit-2 also complements mutant phenotypes in the transformation of nnu mutants in Gibberella zeae (534). The nit-2 mutant is complemented by its homologue area-GF, from Gibberella fujikoroi (2114). There is a possible role in photoreception (1482). Called amr: ammonium regulation (1712).
nit-3 : nitrate nonutilizer-3
IVR. Between met-5 (15%), bd (19%) and pyr-2 (2%; 9%) (1185, 1546, 1950).
Cloned and sequenced: Swissprot NIA_NEUCR, EMBL/Genbank X61303, PIR S16292, S34796, S37298, GenBank NCNIT3.
Structural gene for NADPH nitrate reductase (EC 1.6.6.3) (46). Cannot use nitrate as the nitrogen source, but does use nitrite, ammonia, hypoxanthine, or amino acids (46, 1948) (Fig. 49). Allele 14789 apparently produces an altered enzyme that cannot catalyze the whole electron transport sequence from NADPH to NO3-, but can catalyze the terminal portion of this sequence if a suitable electron donor (reduced viologen dye) is provided (1948). Molecular analyses of mutants (1494). Site-specific mutagenesis of the flavin-binding domain (750) and the heme-binding domain (1495). Nonsense (amber) mutations restorable by ssu-1 (1610). Nuclease-hypersensitive sites in the promoter that depend on NIT-2 and NIT-4 (229). NIT-2 and NIT-4 binding sites in the promoter (375, 2053). For a model of the interaction of nit-1 and nit-3 gene products, see ref. (385). For regulation, see Fig. 50 and refs. (442), (691), (1948), and (1950).
nit-4 : nitrate nonutilizer-4
IVR. Between pyr-1 (1%; 6%) and pan-1 (6%; 27%). Probably right of col-4 (2%) (185, 1546, 1950).
Cloned and sequenced: Swissprot NIT4_NEUCR, EMBL/GenBank M80368, PIR A41696, S20033, EMBL NCNIT4A, GenBank NEUNIT4X.
Structural gene for nitrogen assimilation transcription factor, regulator for induction by nitrate of nitrate reductase and nitrite reductase (2101, 2288-2290), turns on expression of nit-3 and nit-6 (Fig. 50). Cannot use nitrate or nitrite as the nitrogen source, but does use ammonia, and amino acids (185). Nuclease hypersensitive sites in NIT-3 that depend on NIT-2 and NIT-4 (229). NIT-4 binding sites in nit-3 promoter characterized (375, 685). NIT-4 transcriptional activation domain defined (624). Interaction of NIT-2 and NIT-4 is necessary for strong activation of gene expression (625). Complements the Aspergillus nidulans nirA1 mutation (858). The original nit-4 allele was discovered in a wild isolate of Neurospora intermedia from Borneo and was introgressed into Neurospora crassa (185). Allele nr15, called nit-5 (1950), is phenotypically identical to other nit-4 alleles and fails to complement or recombine (0 prototrophs/2080 progeny) (2101).
nit-5 : nitrate nonutilizer-5
Allelic with nit-4.
nit-6 : nitrate nonutilizer-6
VIL. Between chol-2 (6%), T(OY350) and ser-6 (11%), ad-8 (11%, 17%) (1172, 1582, 2277).
Cloned and sequenced: Swissprot NIR_NEUCR, EMBL/GenBank L07391, PIR A49848, EMBL NCNIT6X, GenBank NEUNIT6X, EST SC3H7; pMOcosX clones X4:11A and X6:9F; Orbach-Sachs clone X4A11, X6F9.
Unable to use nitrate or nitrite as the nitrogen source (353). Structural gene for NAD(P)H-nitrite reductase (EC 1.6.6.4) (353, 612) (Figs. 49 and 50), which is subject to positive nitrogen metabolite repression (354). Induced by nitrite (385). Affected by the nit-2 and nmr regulatory genes (1649, 2091). Potential NIT-4-binding sites in promoter (375, 685). Used to study the repression of nitrate reductase (44) and the nonenzymatic reduction of nitrate (353). Directed mutagenesis used to examine the structure-function relationship (395).
nit-7 : nitrate nonutilizer-7
IIIR. Linked to un-17 (0/63) (1546).
Cannot use nitrate or hypoxanthine as the nitrogen source. Resembles nit-1, nit-8, and nit-9 in affecting the molybdenum-containing cofactor common to nitrate reductase and xanthine dehydrogenase (863, 2101, 2102) (Figs. 8 and 49).
nit-8 : nitrate nonutilizer-8
IR. Linked to cr-1 (0/61). Between nic-2 (10%) and thi-1 (3%) (1546).
Cannot use nitrate or hypoxanthine as the nitrogen source. Lacks the molybdenum cofactor for nitrate reductase and xanthine dehydrogenase (863, 2101, 2102) ) (Figs. 8 and 49).
nit-9 : nitrate nonutilizer-9
IVR. Right of nit-4 (9%). Linked to nit-3 (35%; 38%) (2101).
Cloned (563).
Cannot use nitrate or hypoxanthine as the nitrogen source. Lacks the molybdenum cofactor for nitrate reductase and xanthine dehydrogenase (Figs. 8 and 49). A complex locus with three complementation groups, comparable to cnxABC of Aspergillus nidulans (863, 2101, 2102). nit-9 alleles of groups A and B have been cloned (563).
nmr : nitrogen metabolite regulation
VR. Between am (3%; 7%) and gln-1 (4%; 10%) (2100).
Cloned and sequenced: Swissprot NMR_NEUCR, EMBL/GenBank S64286, PIR S11910.
Structural gene for a negative-acting nitrogen regulator (2282) Figs 49 and 50. Synthesis of nitrate reductase is derepressed on ammonium, glutamate, or glutamine. Hypostatic to nit-2 and nit-4. Prototrophic. Isolated and scored by sensitivity to chlorate in the presence of glutamine (568, 2100, 2282). Mutant MS5 (1649) is an allele; a complementing cosmid also complements nit-2 defects (1949). Allele ms5 is different than glnr (269). Levels of nitrate reductase, nitrite reductase, histidase, and acetamidase are elevated in the presence of glutamine and the respective enzyme inducer. The NMR protein binds to two distinct regions of the positive-acting NIT-2 protein, and these interactions are required to establish nitrogen catabolite repression (1527, 2250). Called nmr-1, MS5.
NO : Nucleolus organizer
See NOR.
nop-1 : new opsin-1
VIIL. Linked to ars-1 (2T/18 asci), nic-3 (3T/18 asci) (180).
Cloned and sequenced: EMBL/GenBank AF135863.
Encodes the first opsin identified in fungi or eukaryotic microorganisms, with sequence homology to opsins in animals and the archaea. Expression is stimulated by light and is highest under conditions favoring conidiation. NOP-1 is capable of binding and photocycling retinal (180, 181).
NOR : Nucleolus Organizer Region
VL. At Tip-VL between the terminal sat (118, 1598) and T(AR30), dgr-1, caf-1 (>8%), T(AR33), lys-1 (30%) (118, 1598-1601, 1617). Breakpoints of In(UK2-y), T(NM169), T(ALS169), T(ALS182), T(AR190), and T(OY321) are in the NOR (1578).
Contains rDNA, components of which have been cloned and sequenced. See rDNA. 5.8S sequence: EMBL/GenBank X02447, M10692, NCRRN58S, EMBL NCRGB, GenBank NEURGB, 17S sequence: EMBL/GenBank X04971, GenBank NCRRNAS, 18S sequence (partial): EMBL/GenBank M11033, EMBL NCRRE, GenBank NEURRE, 25S sequence: GenBank X01373; ITS sequence EMBL/GenBank M13906, EMBL NCRGITR, Genbank NEURGITR, 28S sequence (partial): EMBL/GenBank M38154, EMBL NCRRNA1; Orbach-Sachs clone X15E04.
The nucleolus is seen at pachytene to be at one end of the longest chromosome (1307). A dotlike terminal satellite is appended in some strains (see sat). Genes specifying 5.8S, 17S, and 25S ribosomal RNA (but not 5S) are located in the NOR in a series of ~150 tandem repeats (rDNA), each ~9.3 kb long (423, 672). The repeats are separated by nontranscribed spacers (423, 672). Changes in rDNA repeat number occur premeiotically as a result of crossing over between sister chromatids (262-264). The rDNA repeats are presumably located in the attenuated threads seen extending through the nucleolus when acriflavine-stained pachytene chromosomes are examined by fluorescence microscopy (1601, 1672). Nine of eleven chromosome rearrangements that appear genetically to have a terminal breakpoint in VL are in fact physically nonterminal, with a segment of rDNA repeats from the NOR exchanged reciprocally for the terminal segment of another chromosome arm (1590, 1601). T(OY321) divides the NOR into two portions, each of which retains the ability to form a nucleolus (1599). When crossed with normal sequence, T(AR33) produces duplication progeny having two copies of the NOR (1598). These undergo demagnification in such a way that different nontranscribed spacer sequences of both parental types are retained (1733, 1734). Genes specifying 5S ribosomal RNA are not included in the rDNA repeat unit but are located elsewhere in the genome as dispersed single genes flanked by heterogeneous sequences (672, 1893; see Fsr). For research applications of the NOR, see refs. (1578) and (1601).
npd : nitropropane dioxygenase
Unmapped.
Cloned and sequenced: Swissprot 2NPD_NEUCR, EMBL./GenBank U22530.
Structural gene for 2-nitropropane dioxygenase (EC 1.13.11.32) (2063).
nrc-1 : nonrepressor of conidiation-1
Unmapped.
Cloned and sequenced: EMBL/GenBank AF034090; Orbach-Sachs clones X15A05, G8E05, G11G02, G15C05, G16C02.
Encodes a protein kinase homologous to Saccharomyces cerevisiae STE11, which is a MEK kinase (1093). Cannot repress conidiation. Female sterile. Ascospores containing the nrc-1 mutation have an autonomously expressed “flattened” morphology and fail to germinate. Obtained by insertional mutagenesis and identified by altered expression of a ccg-1::tyrosinase reporter gene (1093). The gene was cloned independently by PCR with degenerate primers encoding MEKK-conserved domains, was assigned to Orbach-Sachs clones G05:C06, G16:B08, G16:G02, and was provisionally called mik-2 (1267). The homologs in Saccharomyces and S. pombe are involved in mating-type determination. Proteins of this type phosphorylate MEK proteins in other organisms (1267).
nrc-2 : nonrepressor of conidiation-2
IIR. Linked to Fsr-55. Near cya-4 (676).
Cloned and sequenced: EMBL/GenBank AF034260; Orbach-Sachs clone X7E9.
Encodes a protein kinase homologous to Saccharomyces cerevisiae KIN82 and YNO47W, which are serine-threonine protein kinases. The mutant cannot repress conidiation. Conidia that are produced have a separation defect. Female sterile. Identified by detecting the altered expression of a ccg-1::tyrosinase reporter gene (1093).
nt : nicotinic acid or tryptophan
VIIR. Between arg-10 (2%; 12%) and sk (7%; 18%) (1548).
Uses nicotinic acid. May respond also to tryptophan, phenylalanine, tyrosine, quinic acid, nicotinic acid precursors, and/or tryptophan precursors, depending on genetic background (847, 1474). Best supplemented with nicotinamide and scored as a nic mutant. Probably deficient in tryptophan pyrrolase (tryptophan 2,3-dioxygenase) (EC 1.13.11.11) (Fig. 48), but direct evidence is lacking because tryptophan oxygenase cannot be assayed (702). Kynurenine formamidase levels are normal (702). Pyridine nucleotide levels (234).
ntf2 : nuclear transport factor 2
Unmapped.
Cloned and sequenced: Swissprot NTF2_NEUCR, EMBL/GenBank Y13237, NCNUCTF2.
Homolog of the Saccharomyces gene encoding nuclear transport factor 2 (NTF2) (1342).
nuc : nuclease
Mutants that lack nucleases and phosphatases have been obtained in several ways: by selecting mutants unable to grow on one or more types of nucleic acid (double-stranded DNA, denatured DNA or RNA) as the sole source of phosphorus (659, 850, 967), by selecting mutants with loss or reduction in the size of a halo around the colony from which nucleic acid in the medium has been digested (1014), or by selecting mutants that lack relevant enzyme activity detected with a chromogenic substrate (747, 848, 2096). There is also some phenotype overlap with mutagen sensitivity (see uvs-3 and uvs-6). nuc-2, nuc-3, nuc-4, nuc-5, nuc-6, nuc-7, nuh-5, and nuh-9 are all located on linkage group IIR; it is uncertain whether each of these represents a different locus.
nuc-1 : nuclease-l
IR. Between his-2 (1%), T(AR173)R and met-10, lys-4 (l%) (967, 1022, 1331).
Cloned and sequenced: Swissprot NUC1_NEUCR, EMBL/GenBank M37700, PIR A36378, EMBL NCNUC1, GenBank NEUNUC1.
Structural gene for a transcriptional activator of phosphorus acquisition genes (1022) (Fig. 51). The NUC-1 positive regulatory protein controls the expression of several unlinked target genes involved in phosphorus acquisition and metabolism. It has a helix-loop-helix motif near its C-terminus, which binds to a CACGTG target in the upstream sequences of target genes, e.g., pho-2 and pho-4. The loop between the helices contains an atypical zipper domain. Helix I is required for target DNA binding, whereas the zipper and helix II are required for dimerization (1541). The mutant is defective in the production of repressible alkaline and acid phosphatases (1333, 2096), and several nucleases are absent or reduced (848). nuc-1 mutants (other than nuc-1c) are unable to use RNA or DNA as a phosphorus source (850, 967). Scored on low-phosphate medium by a staining reaction with a-naphthyl phosphate plus Diazo Blue B (747, 2096), by failure to grow on minimal medium altered to substitute 0.l g/l RNA or DNA for an inorganic phosphate source (967, 1014), or by failure to grow on low phosphate at pH above 7 (1322). nuc-1 is epistatic to nuc-2 (called pconc), pregc, and pgovc (1325, 1326, 1331, 1333). In duplications, nuc-1c is dominant to nuc-1+, which is dominant to nuc-1. nuc-1c is scored on high-phosphate medium by staining reaction with a-naphthyl phosphate plus Diazo Blue B (747, 2096) or by suppression of the phenotype of nuc-2 on low phosphate at high pH (1331). Used to study phosphate transport (1229). For regulation model, see refs. (1325), (1326), (1331), and (1541).
nuc-2 : nuclease-2
IIR. Between aro-3, T(NM177)L and preg (1% or 2%), pe (4%). Allelic with pcon (0/854) (967, 1157, 1333).
Cloned and sequenced: EMBL/GenBank U51118, EMBL NC51118; EST SM3A10; Orbach-Sachs clone X2:E11.
Structural gene for an ankyrin repeat protein (1540), a component of the phosphate-regulated signal transduction pathway. Unable to use RNA or DNA as the phosphorus source (967). Defective in the production of repressible alkaline and acid phosphatases (1333, 2096). Several nucleases are absent or reduced (848). Interaction with other phosphate regulatory genes (1325). Recessive to nuc+ in partial diploids and heterokaryons (1333). Not defective in nuh function (1014). Scored on low-phosphate medium by a staining reaction with α-naphthyl phosphate plus Diazo Blue B (747, 2096), by failure to grow on minimal medium altered to substitute 0.1 g/l RNA or DNA for the inorganic phosphate source (967, 1014), or by failure to grow on low phosphate at pH above 7 (1322). Used to study phosphate transport (1229). Regulator gene of repressible alkaline phosphatase (1333) and other steps in phosphorus uptake and metabolism (1322, 1325). Used to study phosphate transport (1229). For regulation model, see refs. (1325) and (1331) (Fig. 51).
nuc-3: nuclease-3
IIR. Right of arg-12 (4%) (659).
Unable to use double- or single-stranded DNA (dsDNA, ssDNA) or RNA as the sole phosphorus source. Reduced extracellular nuclease and alkaline phosphatase levels. Nuclease activity on dsDNA is 2-12% that of wild type. Sensitive to UV, MNNG, and MMS. One of five nuclease mutants (nuc-3 to nuc-7) described in refs. (659) and (1355).
nuc-4 : nuclease-4
IIR. Right of arg-12 (12%) (659).
Unable to use dsDNA as the sole phosphorus source. Reduced extracellular nuclease and alkaline phosphatase levels. Sensitive to MMS, but has antimutator activity against UV and MNNG. One of five nuclease mutants (nuc-3 to nuc-7) described in refs. (659) and (1355).
nuc-5 : nuclease-5
IIR. Right of arg-12 (16%) (659).
Unable to use dsDNA as the sole phosphorus source. Reduced extracellular nuclease and alkaline phosphatase levels. Sensitive to MMS, but has antimutator activity against UV and MNNG. One of five nuclease mutants (nuc-3 to nuc-7) described in refs. (659) and (1355).
nuc-6 : nuclease-6
IIR. Right of arg-12 (23%) (659).
Unable to use dsDNA, ssDNA, or RNA as the sole phosphorus source. Reduced extracellular nuclease and alkaline phosphatase levels. Sensitive to UV, MNNG, and MMS. One of five nuclease mutants (nuc-3 to nuc-7) described in refs. (659) and (1355).
nuc-7 : nuclease-7
IIR. Linked to trp-3 (31%). Apparently unlinked to arg-12 (659).
Unable to use dsDNA, ssDNA, or RNA as the sole phosphorus source. Reduced extracellular nuclease and alkaline phosphatase levels. No increased sensitivity to UV, MNNG, or MMS; in fact, has antimutator effect. One of five nuclease mutants (nuc-3 to nuc-7) described in refs. (659) and (1355).
nuh : nuclease halo
Mutants show reduced nuclease halos on sorbose DNA agar plates (DNase test agar) flooded with HCl (1014). Three nuclease activities have been identified in filtrates of Neurospora grown in Vogel’s minimal medium N (2162) + DNA + sorbose (“sorbose + DNA” medium) (667). Two of these are secreted alkaline DNases (A and B), whose activities are also derepressed 200-fold when cells are grown in phosphate-free medium N + DNA + sucrose (1018). DNase A is a single-strand (ss) and double-strand (ds) endonuclease, specific for DNA (ss-and ds-DNase). DNase B is a periplasmic ss-specific exonuclease, active on DNA and RNA, that can also be isolated from conidia (669). The third nuclease activity corresponds to DNase C, which is an intracellular endo-exonuclease found in nuclear, mitochondrial, and vacuolar compartments and which is not secreted but is released into culture medium when cells are grown on sorbose + DNA media (667). This enzyme has ss-specific endonuclease activity with DNA and RNA and exonuclease activity with ds-DNA, optimal at neutral pH. The Neurospora endo-exonuclease is immunochemically related to the product of Escherichia coli recC (668). The endo-exonuclease is implicated in DNA repair and recombination (669). Some of the isolated nuh mutants showed reduced levels of one or more nucleases, as judged by reduced or missing peaks of ss-DNase activity when chromatographed (667) or by reduced endo-exonuclease activity in extracts (1014). The effects of nuh mutations may be indirect (e.g., altered enzymes for processing or activation of precursors may reduce levels of nuclease activities; 666). Reduced halos may also result from mutations causing reduced growth, reduced secretion, or altered membrane properties (1018). One of the nuh mutants, originally called nuh-4 (FK016), is allelic with the repair mutant uvs-3 (1012). Two other repair mutants also make nuclease halos: uvs-6 (ALS35), and mus-9 [FK104, called uvs(FK104)] (1014, 1017).
nuh-1 : nuclease halo-1
IIIR. Between leu-1 (4%) and nuh-2 (1%), trp-1 (11%) (1014).
Deficient in nuclease released on DNase test agar with sorbose, resulting in a reduced halo around each colony. Reduced levels of alkaline, Mg2+-dependent, ss-DNase activities in extracts of mycelia. Complementation is good between nuh-1 (FK001) and the closely linked nuh-2 (FK027). (Possibly a gene cluster?) Not sensitive to UV or MNNG (1014).
nuh-2 : nuclease halo-2
IIIR. Between leu-1 (4%), nuh-1 (1%) and trp-1 (11%) (1014).
Deficient in the nuclease released on DNase test agar with sorbose, resulting in a reduced halo around each colony. Normal ss-DNase activity in extracts of mycelia (1014). Not sensitive to UV or MNNG (1014).
nuh-3 : nuclease halo-3
VR. Between cyh-2 (4%) and al-3 (17%) (1014).
Deficient in nuclease released on DNase test agar with sorbose, resulting in a reduced halo around each colony. Low levels of DNase B (1014), but normal levels of DNases A and C (667). Normal growth. Fertile in homozygous crosses. Not sensitive to UV, MNNG, or MMS (1018). Two alleles, FK003 and FK004. Closely linked (2-5%) to a complementing mutant, nuh(FK023) (1014).
nuh-4 : nuclease halo-4
Mutant FK016 is allelic with uvs-3.
nuh-5 : nuclease halo-5
IIR. Linked to trp-3 (30%), close to T(4637) al-1 (1014).
Deficient in nuclease released on DNase test agar, resulting in a reduced halo around each colony. Nuclease activities in extracts are 55% of wild type (1014). Mutant FK005 is closely linked to a partially complementing mutant, nuh(FK029) (1008). (Possibly a complex gene or a gene cluster?)
nuh-6 : nuclease halo-6
IR. Between Cen-I (5%) and nic-2 (4%) (1014).
Deficient in nuclease released on DNase test agar, resulting in a reduced halo around each colony. Levels of tested nucleases in extracts are normal. Not sensitive to mutagens. Growth is normal (1014).
nuh-7 : nuclease halo-7
Unlinked to any mapped nuh gene or to alcoy markers (1018).
Decreases the nuclease released on DNase test agar, resulting in a reduced halo around each colony. Very low ss-DNase and reduced ds-DNase activity in filtrates, even under derepressing conditions. Early growth is normal (up to 16 hrs), but growth is somewhat reduced later (3-5 days), with an increasingly high release of protein into the filtrate. Small amounts of ss- and ds-DNase are secreted early, but not later. Lack of activity is not due to the formation of an inhibitor. Overall intracellular ss-DNase activities (presumably DNase C) are 50-80% of wild type (1018). nuh-7 (FK017) probably is not a nuclease structural mutation, but may be altered in secretion or membrane structure.
nuh-8 : nuclease halo-8
IR. Right of nic-1 (1014, 1018).
Deficient in nuclease released on DNase test agar, resulting in a reduced halo around each colony. Growth and UV survival are normal, as is the enzyme profile of secreted DNases A and B in medium N + sorbose. There is virtually no derepression in phosphate-free medium N + DNA + sucrose (less than 2-fold, compared to the 200-fold normally observed). One allele, (FK018) (1018). Possibly in a cluster with nuh(FKO40) (1008).
nuh-9 : nuclease halo-9
IIR. Linked to arg-5. Left of mus-7, nuc-2 (1018).
Deficient in nuclease released on DNase test agar, resulting in a reduced halo around each colony. Increased growth and release of ss-DNase in sorbose +DNA medium, but growth and levels of induced ss- and ds-DNase are variably reduced in phosphate-free medium N + DNA + sucrose. The mutant is therefore difficult to use for detailed analysis (1018). Moderately sensitive to MMS. Barren in homozygous crosses.
nuh-10 : nuclease halo-10
VIR. Linked to trp-2 (1018).
Deficient in nuclease released on DNase test agar, resulting in a reduced halo around each colony. Mutant FK028 shows reduced growth and a reduced level of DNase A secretion in phosphate-free medium N + DNA + sucrose. Growth is normal, but ss-DNase activity is reduced overall in sorbose + DNA medium (1018). MMS sensitivity is not increased. Fertility is normal.
nuh(19) : nuclease halo (19)
Unlinked to any mapped nuh gene or to alcoy markers (1018).
Deficient in nuclease released on DNase test agar, resulting in a reduced halo around each colony (1018).
nuh(22) : nuclease halo (22)
Unlinked to any mapped nuh gene or to alcoy markers (1018).
Deficient in nuclease released on DNase test agar, resulting in a reduced halo around each colony. Nuclease profiles are normal. Poor fertility when homozygous. Sensitivity to MMS is moderately increased (1018).
nuh(41) : nuclease halo (41)
Unmapped.
Deficient in nuclease released on DNase test agar, resulting in a reduced halo around each colony. Nuclease profiles are normal on sorbose + DNA medium and close to normal under derepressing conditions. Sterile in homozygous crosses. MMS sensitivity is increased slightly (1018).
nuh(42) : nuclease halo (42)
Unmapped.
Deficient in nuclease released on DNase test agar, resulting in a reduced halo around each colony. Single-nuclease levels are reduced in filtrates from cells grown in either sorbose + DNA medium or phosphate-free medium N + DNA + sucrose, whereas growth is reduced only slightly in either medium (1018).
nuo : NADH:ubiquinone oxidoreductase
The mitochondrial NADH:ubiquinone complex (EC 1.6.5.3), known as respiratory complex I, has a total size of approximately 700kDa. Complex I of Neurospora has been imaged at medium resolution (682, 808, 886). It consists of two arms: one membrane-embedded and one peripheral, protruding into the mitochondrial matrix. Within complex I are at least 28 nucleus-encoded and 7 mitochondrion-encoded subunits (2203). Two subcomplexes of subunits from the two sources appear to be assembled separately prior to final assembly of the native complex in the mitochondrial membrane. The nomenclature for the nuclear-encoded genes of the complex (nuo genes) incorporates the size of each polypeptide product in the gene name; when multiple genes specify products of similar size, they are differentiated further with letters (e.g., nuo21.3a, nuo21.3b, and nuo21.3c each encode a different 21.3-kDa subunit). Mitochondrion-encoded subunits include ndh1 (41.5-kDa subunit), ndh2, ndh3, ndh4, ndh4l, ndh5 (80-kDa subunit), and ndh6 (see Appendix 4 for ndh genes). Blocking the synthesis of the mitochondrial subunits with chloramphenicol results in the synthesis of a smaller, functionally different form of complex I (683, 2183). Disruptions of each of the nuclear genes can cause mild to serious assembly defects that eliminate complex I function. Loss of complex I can slow asexual growth and reduce conidiation, but the vegetative phenotypes appear surprisingly mild. In contrast, complex I appears essential for sexual development (554). Reviews (783, 2149, 2203). See also acp-1.
nuo9.3 : NADH:ubiquinone oxidoreductase 9.3
Unmapped.
Cloned and sequenced: Swissprot NI9M_NEUCR, EMBL/GenBank S49807, PIR A44210.
Encodes the NADH:ubiquinone oxidoreductase 9.3-kDa subunit (EC 1.6.5.3) (866).
nuo9.6 : NADH:ubiquinone oxidoreductase 9.6
A synonym of acp-1.
nuo10.5 : NADH:ubiquinone oxidoreductase 10.5
Unmapped.
Cloned and sequenced: Swissprot NI8M_NEUCR, EMBL/GenBank X69929, PIR S30186, GenBank NCMIT.
Encodes the NADH:ubiquinone oxidoreductase 10.5-kDa subunit (EC 1.6.5.3) (552).
nuo12.3 : NADH:ubiquinone oxidoreductase 12.3
IR. Between lys-4 (1T/18 asci) and cys-9 (7T/18 asci) (1447).
Cloned and sequenced: Swissprot NUMM_NEUCR, EMBL/GenBank X68965, NCNUO, PIR S29557, S32568.
Encodes the NADH:ubiquinone oxidoreductase 12.3-kDa subunit (EC 1.6.5.3) (2150). Disruption causes assembly defects (555).
nuo14 : NADH:ubiquinone oxidoreductase 14
Unmapped.
Cloned and sequenced: EMBL/GenBank Z18945, NCUBIOXRI, PIR S47150.
Encodes the NADH:ubiquinone oxidoreductase 14-kDa subunit (EC 1.6.5.3) (1445)
nuo14.8 : NADH:ubiquinone oxidoreductase 14.8
Unmapped.
Cloned and sequenced: Swissprot NB4M_NEUCR, EMBL/GenBank X76344, GenBank NCUO.
Encodes the NADH:ubiquinone oxidoreductase 14.8-kDa subunit (EC 1.6.5.3) (75).
nuo17.8 : NADH:ubiquinone oxidoreductase 17.8
Unmapped.
Cloned and sequenced: Swissprot NURM_NEUCR, EMBL/GenBank X71414, NCNUO178, PIR S35057; EST SM1F6.
Encodes the NADH:ubiquinone oxidoreductase 17.8-kDa subunit (EC 1.6.5.3) (75).
nuo18.3 : NADH:ubiquinone oxidoreductase 18.3
Unmapped.
Cloned and sequenced: EMBL/GenBank X56226, GenBank NCNHU18.
Encodes the NADH:ubiquinone oxidoreductase 18.3-kDa subunit (EC 1.6.5.3) (2200)
nuo19.3 : NADH:ubiquinone oxidoreductase 19.3
VIL. Between gdh-1 (9T/18 asci) and asd-1 (5T/18 asci), Bml (10T/18 asci) (632, 1447).
Cloned and sequenced: EMBL AJ001520, NCIRONSUL.
Encodes the NADH:ubiquinone oxidoreductase 19.3-kDa subunit (EC 1.6.5.3). Homolog of the PSST subunit of bovine complex I (1953).
nuo20.8 : NADH:ubiquinone oxidoreductase 20.8
IL. Between mat (0 or 1T/17 asci) and Cen-I (3T/17 asci) (1447).
Cloned and sequenced: Swissprot NUPM_NEUCR, EMBL/GenBank M55323, PIR A36621, GenBank NEUCOMPI.
Encodes the NADH:ubiquinone oxidoreductase 20.8-kDa subunit (EC 1.6.5.3). Disruption causes assembly defects (438).
nuo20.9 : NADH:ubiquinone oxidoreductase 20.9
Unmapped.
Cloned and sequenced: Swissprot NUXM_NEUCR, EMBL/GenBank X60829, PIR S27171, GenBank NCNADHC1.
Encodes the NADH:ubiquinone oxidoreductase 20.9-kDa subunit (EC 1.6.5.3) (77). Disruption causes an assembly defect (1843).
nuo21 : NADH:ubiquinone oxidoreductase 21
IVR. Linked to Tel-IVR (1T/18 asci), Fsr-4 (1 or 2 T/18 asci) (1447).
Cloned and sequenced: Swissprot NUYM_NEUCR, EMBL/GenBank X78082, GenBank NCNUO21, PIR S17192.
Encodes the NADH:ubiquinone oxidoreductase 21-kDa subunit (EC 1.6.5.3) (76). Erroneously called nuo18 due to a sequencing error that gave a predicted truncated product (2200).
nuo21.3a : NADH:ubiquinone oxidoreductase 21.3a
VR. Left of inl (18%), inv (22%)(41).
Cloned and sequenced: Swissprot NUIM_NEUCR, EMBL/GenBank M32244, GenBank NEUCOM1S.
Encodes the NADH:ubiquinone oxidoreductase 21.3-kDa subunit 21.3a (EC 1.6.5.3). The polypeptide product is found in the peripheral arm of complex I. Referred to as 21.3a in reviews (2149, 2203) and as 21.3b in some primary literature (41, 79). Disruption causes an assembly defect (41).
nuo21.3b : NADH:ubiquinone oxidoreductase 21.3b
Unmapped.
Cloned and sequenced: Swissprot NUJM_NEUCR, EMBL/GenBank X56612, GenBank NCNURI, PIR S14277.
Encodes the NADH:ubiquinone oxidoreductase 21.3-kDa subunit 21.3b (EC 1.6.5.3) (1444). Polypeptide product is found in the membrane arm of complex I. Referred to as 21.3b in reviews (2149, 2203) and as 21.3a in some primary literature (79). Disruption causes an assembly defect (1443).
nuo21.3c : NADH:ubiquinone oxidoreductase 21.3c
VIR. Linked to Tel-VIR (0 or 1T/18 asci) (632, 1447, 2149).
Cloned and sequenced: EMBL/GenBank X95547, GenBank NCMC1.
Encodes ferredoxin-like NADH:ubiquinone oxidoreductase 21.3-kDa subunit 21.3c (EC 1.6.5.3) (553). Disruption causes an assembly defect (2149).
nuo24 : NADH:ubiquinone oxidoreductase 24
VR. Linked to cyh-2 (0T/18 asci) (632, 1447, 2149).
Cloned and sequenced: Swissprot NUHM_NEUCR, EMBL/GenBank X78083, GenBank NCNOU24.
Encodes the NADH:ubiquinone oxidoreductase 24-kDa subunit (EC 1.6.5.3) (76). Disruption causes an assembly defect (2149).
nuo29.9 : NADH:ubiquinone oxidoreductase 29.9
IVR. Between trp-4 (8 or 9 T, 1 NPD/18 asci) and Tel-IVR (8T, 1NPD/18 asci) (1447).
Cloned and sequenced: Swissprot NUFM_NEUCR, EMBL/GenBank X56237, GenBank NCNUO32, PIR S17191.
Encodes the NADH:ubiquinone oxidoreductase 29.9-kDa subunit (EC 1.6.5.3) (2140). Disruption causes an assembly defect (555).
nuo30.4 : NADH:ubiquinone oxidoreductase 30.4
VIL. Linked to Bml, Cen-VI, pan-2 (0T/18 asci) (554, 1447).
Cloned and sequenced: Swissprot NUGM_NEUCR, EMBL A35935, PIR A35935.
Encodes the NADH:ubiquinone oxidoreductase 30.4-kDa subunit (EC 1.6.5.3) (554). Disruption causes an assembly defect (554).
nuo40 : NADH:ubiquinone oxidoreductase 40
Unmapped?
Cloned and sequenced: Swissprot NUEM_NEUCR, EMBL/GenBank X56238, GenBank NCNUO40, PIR S13025.
Encodes the NADH:ubiquinone oxidoreductase 40-kDa subunit (EC 1.6.5.3). Disruption causes an assembly defect (1843); the complex lacking this subunit does not contain tightly bound NADPH (1842).
nuo41.5 : NADH:ubiquinone oxidoreductase 41.5
Not a nuclear gene. See mitochondrial gene ndh (Appendix 4).
nuo49 : NADH:ubiquinone oxidoreductase 49
Unmapped
Cloned and sequenced: Swissprot NUCM_NEUCR, EMBL/GenBank X54508, GenBank NCNUO49, PIR S13801.
Encodes the NADH:ubiquinone oxidoreductase 49-kDa subunit (EC 1.6.5.3). Disruption causes an assembly defect (1843).
nuo51 : NADH:ubiquinone oxidoreductase 51
Unmapped
Cloned and sequenced: Swissprot NUBM_NEUCR, EMBL/GenBank X56227, PIR S17663, GenBank NCNDU51.
Encodes the NADH:ubiquinone oxidoreductase 51-kDa subunit (EC 1.6.5.3), which is the NADH-binding subunit. Disruption causes an assembly defect (616).
nuo78 : NADH:ubiquinone oxidoreductase 78
IIR. Linked to preg (0T/15 asci) (1447).
Cloned and sequenced: Swissprot NUAM_NEUCR, EMBL/GenBank X57602, L36813, PIR S17664, S59926, GenBank NCNDU78, NEUNDS.
Encodes the NADH:ubiquinone oxidoreductase 78-kDa subunit (EC 1.6.5.3). Disruption by sheltered RIP causes an assembly defect (840).
nuo80 : NADH:ubiquinone reductase 80
Not a nuclear gene. See mitochondrial gene ndh5 (Appendix 4).
Nystatin Resistance
See erg.
nac : adenylate cyclase
Allelic with cr-1.
Return to the 2000 Neurospora compendium main page