pa : pale
IR. Between cr-1 (10%) and dir (37%) (1199, 1200).
Conidia are sparse, clumped, and pale. Photograph (1200). Stock lost. (Possibly wc-2?)
pab : para-aminobenzoic acid
p-Aminobenzoate is produced from chorismate in the aromatic amino acid biosynthetic pathway (Fig. 13). The holoenzyme, p-aminobenzoate synthase (EC 4.1.3.-), has two components, ADC synthase (I) and glutamine amidotransferase (II). Typically these are encoded by two different genes. Which of the two pab genes in Neurospora specifies which activity is uncertain.
pab-1 : para-aminobenzoic acid-1
VR. Between inl (1%; 10%) and trp-5, met-3 (1% or 2%) (673, 2014).
Specifies a component of p-aminobenzoate synthase (EC 4.1.3.-) (Fig. 13). Requires p-aminobenzoic acid (2058). Apparently cannot use folate (mono- or triglutamate) (2293).
pab-2 : para-aminobenzoic acid-2
VR. Linked to ro-4 (0/407). Between ad-7 (8%), T(EB4)R and mus-11 (7%), inv (3%), asn (1%; 15%) (321, 1578, 1592, 1603, 1794, 1828, 2014).
Specifies a component of p-aminobenzoate synthase (EC 4.1.3.-) (Fig. 13). Requires p-aminobenzoic acid (2304). Mutant 71301, called pab-3 (2304), was shown to be allelic (550).
pab-3 : para-aminobenzoic acid-3
Allelic with pab-2.
Pad-1 : Paddle-1
IIL. Between het-c and pyr-4 (1%); restriction fragment G22:H5 with het-c, X7:F1 with pyr-4 (1109).
Cloned and sequenced: EMBL/GenBank AF130355.
Sequence characteristics resemble those of heterogeneous nuclear RNA-binding proteins and mRNA- splicing factors. Pad-1 function is essential for sexual development and vegetative growth. Partially functional mutants obtained by RIP show ascus-dominant defects in ascus and ascospore formation. Some asci are swollen and paddle-shaped, with no apical pore and no ascospores. Nuclear divisions and spindle orientation are abnormal in other asci that are not paddle-shaped. Two, four, or eight ascospores may be cut out in these asci, but the ascospores are inviable. Vegetative growth is severely defective. The growth defect is recessive in heterokaryons (1109).
pan-1 : pantothenic acid-1
IVR. Between ad-6 (1% or 2%) and cot-1 (2% or 3%) (1255, 1369, 1582). cel-1 (1%), col-1 (0T/47 asci), int (0/50), pho-3 (<1%), ro-1 (0/394), and thi-5 (1%) also are closely linked.
Cloned: pSV50 clone 31:9C, Orbach-Sachs clones X3B04, X11A07, X18G05, G19F10.
Requires intact pantothenic acid for growth under standard conditions. Able to synthesize both precursors, b-alanine and pantoyllactone (2059). The ability to synthesize pantothenic acid from b-alanine + pantoyllactone is demonstrable in vitro but not in vivo unless cultures are aerated (2173, 2175, 2176). Unlike pan-2, pan-1 has no effect on ascospore ripening in heterozygous crosses. Called group A. For alleles see ref. (296).
pan-2 : pantothenic acid-2
VIR. Between rib-1 (<1%; 3%) and del (6%), trp-2 (11%) (299, 300, 302, 1582, 1603).
Cloned and partially sequenced: Orbach-Sachs clone X6D10 (572, 1738a).
Encodes ketopantoate hydroxymethyltransferase (EC 2.1.2.11) (1738a). Unable to convert ketovaline to ketopantoic acid (296, 299, 300). Used in major studies of intralocus recombination and complementation (299-302). pan-2 ascospores remain white or pale if the crossing medium is not supplemented, even when the protoperithecial parent is pan-2+. Asci in which gene conversion has occurred at pan-2 thus can be recognized and isolated (2086, 2087); photographs (2086). For good recovery of pan-2 progeny, crossing media should be supplemented with pantothenic acid (10 mg/ml), even when the protoperithecial parent is pan+. Called group B.
pat : patch
IL. Linked to mat, probably to the right (1988).
Growth and conidiation occur in patches, and in a cyclic pattern under certain conditions (1988). Initially found in a pro-1 21863 strain, but the pro-1 mutation is not necessary for the expression of patch. The original patch isolates were all sorbose-resistant (1988). A sorbose-resistant derivative that does not express the patch phenotype is called sor-4 (1592). It is not clear whether pat and sor-4 are separate genes, or whether patch is not scorable in the absence of modifiers present in the parent stock [see p. 267 of ref. (1592)]. The original patch strain was used for the first demonstration of a circadian rhythm in fungi (223, 1628).
pcna : proliferating cell nuclear antigen
VL. Linked to rDNA (0T/18 asci) (1091).
Cloned and sequenced: Orbach-Sachs cosmid X2-H8 (1091).
Encodes the Neurospora homolog of proliferating cell nuclear antigen (PCNA), first identified in mammals and known to be a ring-shaped homotrimer necessary for processive DNA synthesis in eukaryotes (1091).
pcon : phosphatase control
Allelic with nuc-2.
pdc-1 : pyruvate decarboxylase-1
Allelic with cfp.
pde-1 : GTP-regulated cyclic phosphodiesterase-1
Unmapped. Perhaps in linkage group II (844).
Affects GTP-regulated cyclic phosphodiesterase (845).
pdx : pyridoxine
Mutants at two closely linked loci require pyridoxine, pyridoxal, or pyridoxamine for growth (1657, 1660, 1661). The first nutritional mutant ever found was a pyridoxine auxotroph (135). Pyridoxine mutants provided the first proved example of gene conversion (1663). Early pdx mutants were called pdx-1 except for isolate 44204, which was called pdx-2 on the basis of one crossover (see 1663). Mutant 44204 was subsequently reclassified as pdx-1 on the basis of noncomplementation with certain of the alleles then designated pdx-1. The existence of two genes has now been reestablished, placing all pH-sensitive pdx mutants into pdx-2 and non-pH-sensitive mutants into pdx-1 (507a). Isolates originally called pdxp and considered alleles of pdx-1 are pH-sensitive, growing without supplement on medium containing ammonium ions above pH 6 (2006). These now appear to be pdx-2 alleles (570a). The two pdx loci are adjacent, and divergently transcribed. There is complementation between mutants (1660, 1661) and intralocus recombination (1663). Many alleles are leaky. Scoring is sharpened by the addition of 100 mg desoxypyridoxine per liter (1660). Conidia are subject to death by unbalanced growth on minimal medium (2010). A yellow pigment is excreted under certain conditions by the double mutant pdx; En(pdx); see En(pdx).
pdx-1 : pyridoxine-1
IVR. Between pyr-1 (1%; 10%) and T(S1229)L, pt (2%) (87, 111, 1369, 1580, 570a).
Cloned and sequenced: Orbach-Sachs cosmid G6G8 (1441a, 1446, 570a).
Pyridoxine-requiring isolates originally classified as type a (1660, 1661, 570a). Not pH-sensitive. Shows strong sequence identity to the highly conserved gene snz in Saccharomyces (224) and to SOR1 (singlet oxygen resistance) in Cercospora nicotianae. SOR1 function is essential for pyridoxine synthesis both in Cercospora and in Aspergillus flavus (599). Linkage data place pdx-1 and snz in the same short region in Neurospora (1447). Five sequenced pdx-1 mutants are altered in the Neurospora gene previously identified as snz (1441a). Thus the Neurospora snz gene is allelic with pdx-1.
pdx-2 : pyridoxine-2
IVR. Between pyr-1 (1%; 10%) and T(S1229)L, pt (2%). Near pdx-1, probably to the left (87, 111, 1369, 1580, 570a).
Cloned and sequenced: Orbach-Sachs cosmid G6G8 (1441a, 1446, 570a).
Pyridoxine-requiring isolates originally classified as types b and g (1660, 1661, 570a). pH-sensitive. Homologous with sno1 in Saccharomyces (599, 570a).
pe : peach
IIR. Between nuc-2 (4%) and arg-12 (1%; 5%) (1157, 1592).
Distinctive morphology (102, 1203). Peach-colored conidia. Short hyphae are formed more uniformly than in wild type, as a lawn close to the agar surface. Added arginine increases macroconidiation and tends to obscure scoring of pe at 25ºC but not at 39ºC. The single mutant pe produces both macro- and microconidia. The double mutant pe fl produces abundant gray microconidia and no macroconidia (102, 1389) (see fl). See col-1, col-4, and refs. (781) and (782) for interactions with other mutants. Called pem or m in some contexts.
pen-1 : perithecial neck-1
VIIL. Linked to csp-2 (4%) (1593).
When the pen-1 mutant is used as female parent, perithecia lack necks (beaks) and ascospores are not shot, although a few ascospores may be formed. When pen-1 is used to fertilize a pen+ strain, perithecial beaks are normal and crosses are fully fertile (518, 1593).
pep : processing enhancing protein
Unmapped.
Cloned and sequenced: Swissprot MPP2_NEUCR, EMBL/GenBank M20928, PIR A29881, S03968, GenBank NEUPEP.
Encodes the 52-kDa subunit of mitochondrial-processing peptidase (EC 3.4.24.34). PEP cooperates with MPP (mitochondrial-processing peptidase, encoded by mpp) (1823) in the proteolytic cleavage of matrix-targeting sequences from nuclear-encoded mitochondrial precursor proteins (860).
pep4 : PEP4 homolog
VIIR. Linked to for, frq (0T/18 asci) (1447). On a common cosmid with frq (2144).
Cloned and sequenced: Swissprot CARP_NEUCR, EMBL/GenBank U36471.
Structural gene for vacuolar proteinase A, homologous with Saccharomyces cerevisiae PEP4 (E.C. 3.4.23.-) (2145). Contrary to the name, the substrates of Neurospora crassa PEP4 are proteins rather than peptides (204).
per-1 : perithecial-1
VR. Between at (8%; 14%), asp (16%; 26%) and ilv-1 (4%) (919, 1582), ts (25%) (1002).
Perithecial walls fail to blacken when per-1 is the female parent (Fig. 37), regardless of the genotype of the fertilizing parent (918, 919, 1002). Alleles are of two types (918): In Type I (e.g., alleles PBJ1, ABT8, AR174), per-1 ascospores are unpigmented and perithecial walls are completely devoid of black pigment. In Type II (e.g., alleles 29-278, 29-281, UG1837), per-1 ascospores are the normal black and the initially unpigmented perithecial walls become brownish as they age. Black pigment develops in a ring around the ostiole of type II perithecia, but is pale or lacking in Type I (918). Unlike the perithecial wall, the ascospore trait shows no maternal effect. White per-1 ascospores (type I) germinate without heat shock and are usually killed by hypochlorite or by the 30-min, 60ºC treatment normally used to activate ascospores (918, 1002). Expression is completely autonomous in ascospores [photographs in ref. (999)] and at least partially so in the perithecial wall (998, 999, 1002). Mosaic perithecia from heterokaryons have been used for a clonal analysis of perithecial development (998, 1002). Used to show that perithecial walls are capable of making orange carotenoids (1569). Used in an unsuccessful search for variegated-type position effects (1001). Beaks of perithecia homozygous for allele PBJl (type I) are abnormal, and ascospores are not shot properly (1673). Type I alleles were initially called sw: snow white (1002).
Perithecial Development Mutants
See fs, ff, fmf, mb, mei, pen, per, sdv.
Permease
See Transport.
pex2 : peroxidase assembly factor 2
IL. Near mat. Between cyt-21 and mei-3 (921).
Cloned and sequenced: pSV50 cosmid 19:3E (921).
Homologous to human pex2 and to Podospora car-1 (161), which was used for cloning the Neurospora gene. Encodes a putative peroxisome assembly factor with homology to PAF1, which is implicated in the lethal Zellweger Syndrome in humans. In Neurospora, disruption of pex2 by RIP probably impairs peroxisome function. The mutant is resistant to 3-AT. Growth is poor in the presence of oleic acid. Sexual development is delayed when pex2 is used as female parent (921).
pf : puff
IVR. Right of pyr-2 (2%). Linked to rug (3%) (1585, 1928).
Spreading colonial growth (1585).
pfa-1 : polyunsaturated fatty acids-1
VII. Linked to csp-2 (35%) (756).
Involved in fatty acid desaturation, with reduced levels of linoleate and linolenate (753, 756) (Fig. 17).
pfa-2 : polyunsaturated fatty acids-2
IV or V. Linked to T(R2355), cot-1 (32%) in cross to alcoy (756).
Involved in fatty acid desaturation, with reduced levels of linoleate and linolenate (753, 756) (Fig. 17).
pfa-3 : polyunsaturated fatty acids-3
IV or V. Linked to T(R235), cot-1 (29%) in cross to alcoy (756).
Involved in fatty acid biosynthesis, with an indirect effect on the desaturation of linolenate. Accumulates free fatty acids (756) (Fig. 17). [Free fatty acids are also accumulated by mitochondrial mutant [mi-1] (737).]
pfa-4 : polyunsaturated fatty acids-4
VII. Linked to csp-2 (33%) (756).
Involved in fatty acid biosynthesis, with an indirect effect on the desaturation of linolenate (753, 756).
pfa-5 : polyunsaturated fatty acids-5
I or II. Linked to T(4637) al-1 (7%) in cross to alcoy (756).
Involved in fatty acid biosynthesis, with an indirect effect on the desaturation of linolenate (753, 756).
pgk : phosphoglycerate kinase
Unmapped.
Cloned and sequenced: Swissprot PGK_NEUCR, EMBL/GenBank X56512, GenBank NCPGKR, PIR A56616; EST NC3D6.
Structural gene for phosphoglycerate kinase (EC 2.7.2.3) (78).
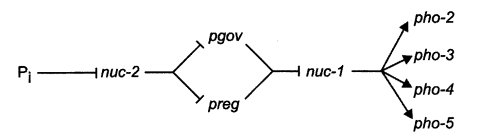
pgov : phosphorus governance
IIIR. Linked to tyr-1 (1%; 4%), probably to the right (1322).
Regulatory gene in phosphorus uptake and metabolism (1325, 1326, 1331) (Fig. 51). Probably specifies a cyclin-dependent kinase. pgov corresponds to a gene cloned by P. Margolis and called mdk-1. pgovc is similar to pregc phenotypically (1322, 1325). Isolated in preg+/preg+ partial diploids. Mutants were obtained first as a suppressor of nuc-2 and then by insertional mutagenesis. pgov allele c-5 is largely or completely recessive in duplications from T(D305) (1322). Scored on high-phosphate medium by staining reaction with a-naphthyl phosphate plus Diazo Blue B (747, 2096).
phe-1 : phenylalanine-1
IL. Between suc (<1%), In(H4250)L and ad-5 (914, 1592).
Originally reported to grow on phenylalanine, other aromatic amino acids, leucine, or ethyl acetoacetate, with phenylalanine being the most effective; several other acids gave smaller responses (104). Utilization of phenylalanine and other compounds varies for different isolates and on different carbon sources; glycerol or ribose are preferable to sucrose (992-994, 1464). Allele NM160 does not use phenylalanine, but grows well on tyrosine or leucine (510, 1592), at least with the strains and carbon source used. phe-1 is inhibited by basic amino acids on low phenylalanine or on leucine (104, 992). Growth on b-labeled leucine or b-labeled phenylalanine showed that neither compound is converted to the other (101). The mutant has a 19-hr circadian period, even on high levels of phenylalanine. The high levels of phenylalanine required for growth allow the synthesis of sterols by an alternate catabolic pathway, as evidenced by patterns of incorporation of radioactive phenylalanine and acetate into sterols [S. Brody, unpublished, cited in ref. (1130)]. Called phen-1. Allele NM160 called tyr(NM160) (601).
phe-2 : phenylalanine-2
IIIR. Linked to vel (1%). Between T(D305)L and tyr-1 (2%; 4%) (87, 601, 1581).
Lacks prephenic dehydratase (EC 4.2.1.51) (87, 601) (Fig. 13). The requirement is very leaky. Grows extensively and is treacherous to score by growth on minimal vs supplement, but can be scored reliably by blue fluorescence under long-wave UV after growth on minimal medium without phenylalanine (1592). The appearance of phenylalanine in the culture medium (245) is due to spontaneous conversion of accumulated pretyrosine (988). Called phen-2. Allele Y16329 called phen-3 (601).
phe-3 : phenylalanine-3
Allelic with phe-2.
phen : phenylalanine
Symbol changed to phe.
pho-1 : phosphorus-1
II? 20% wild-type recombinants with nuc-2. Independent of nuc-1.
Activity of the repressible alkaline phosphatase is low (Fig. 51). Complements nuc-1 and nuc-2 in heterokaryons. Stains pale red on low-phosphate medium with a-naphthyl phosphate + Diazo Blue B (2096).

FIGURE 51Regulation of genes concerned with phosphorus acquisition in N. crassa. The nuc-2+ gene encodes an ankyrin-repeat protein, pgov+ probably encodes a cyclin-dependent kinase, preg+ encodes a cyclin, and nuc-1+ encodes a helix-turn-helix transcriptional activator. Redrawn from ref. (1326 ), with permission. Copyright American Society of Plant Physiologists.
pho-2 : phosphorus-2
VR. Between his-1 (3%) and inl (4%) (747, 1322).
Cloned and sequenced: EMBL/GenBank L27993, GenBank NEUALPH; pSV50 clone 23:1A.
Structural gene for Pi-repressible alkaline phosphatase (EC 3.1.3.1) (747, 1158, 1455). Scored on low- phosphate medium by a staining reaction with a-naphthyl phosphate plus Diazo Blue B (747, 2096). Encodes multiple forms (10 or more) of the enzyme from the same coding sequence (2081) (Fig. 51).
pho-3 : phosphorus-3
IVR. Linked to pan-1 (<1%) (1455).
Structural gene for repressible acid phosphatase, with phosphodiesterase activity (1455) (Fig. 51). Codominant in heterozygous pho-3/pho-3+ duplications. Scored on low-phosphate medium by a staining reaction with bisnitrophenyl phosphate (1455).
pho-4 : phosphate-4
VIIL. Linked to nic-3 (2-6%; 1 or 2T/18 asci) (205, 1447).
Cloned and sequenced: Swissprot PHO4_NEUCR, EMBL/GenBank M31364, PIR JQ0116, GenBank NEUPHO4A; pSV50 clone 24:8F, Orbach-Sachs clones X20D10, G3D12.
Structural gene for phosphate-repressible phosphate permease (1259) (Fig. 51). Resistant to vanadate (0.1-1.0 mM) in Vogel's minimal medium with 0.15 mM phosphate and 20 mM HEPES buffer, adjusted to pH 7.5 (204). Constitutive mutation Ab(RLM01) is inseparable from pho-4 (1578). Called van.
pho-5 : phosphorus-5
IVR. Between pyr-1 (5%; 2T/18 asci) and trp-4 (4T/18 asci), cot-1 (15%) (1447, 2147).
Cloned and sequenced: EMBL/GenBank L36127, GenBank NEURHAPPA; Orbach-Sachs clone G16E10.
Structural gene for repressible high-affinity phosphate permease (Fig. 51). Obtained as a suppressor in nuc-1; pho-4. Constitutive mutations T(RLM02), T(RLM04), T(RLM06), T(RLM08), T(RLM09), and T(RLM11) are inseparable from pho-5 (1578).
phr : photoreactivation deficient
IR. Right of os-1 (15%). Linked to con-8 (0/18), nic-1 (4/18) (1908).
Cloned and sequenced: Swissprot PHR_NEUCR, EMBL/GenBank X58713, GenBank NCPHR, PIR S18667.
Epistasis group Phr (Table 3). Structural gene for deoxyribodipyrimidine photolyase (DNA photolyase) (EC 4.1.99.3) (600, 2252). The RIP-inactivated phr mutant is defective in photolyase activity for cyclobutane-pyrimidine dimers and TC(6-4)-photoproducts caused by UV irradiation. Dark repair is not affected. Neurospora photolyase is not a blue light receptor (1908). Called uve-1.
pi : pile
IIL. Linked to ro-7 (0/75). Between T(AR30) (28%) and cys-3 (4%) (1173, 1578, 1592).
Slow, spreading mycelial growth. No conidia are formed (1592). Scanning EM photograph (1955): few major constriction chains. pi B101 grows better on minimal medium than on complex complete medium. col-10 R2438 is a putative allele (1852). Because of growth rate, stability, and ease of handling, pi B101 is preferable to col-10 R2438 as a marker. ro-7, in the same region, is preferable to both (1582).
pk : peak
VR. Between met-3 (1%) and T(EB4)L, cot-2 (8%), cl (2%) (569, 1582, 1603, 2014).
Growth on an agar surface is initially colonial and flat. A mass of aerial hyphae is then sent up, which conidiates profusely (1548). Somewhat similar in morphology to sn, cum, sp, and cot-4 at 25ºC, but distinguishable. Hyphae branch dichotomously (1405, 1548). Increased activity of L-glutamine:D-fructose-6-phosphate amidotransferase was observed in crude extracts of one pk strain, but not in nine others; increased activity for this enzyme also was found in cl and in four other nonallelic morphological mutants (1758). Hexoseaminoglycan consists of a single component on medium without sorbose, in contrast to two components in wild type (1960). Antigenic surface mucopolyoside (532). Cell-wall analysis and photograph, allele B6 (507) and allele C-1810-1 (287). Cell-wall enzymes (633). Effect of carbon source (531). One observation suggested a functional interaction with cl (1965), but substantial crossing-over frequencies and recovery of the double mutant pk cl indicated that the loci are distinct (569). The gene was named for the vegetative mutant phenotype, which is recessive. Several alleles were called bis (1592). The sexual phase is also affected. Asci are thin-walled, bulbous, and nonlinear in homozygous pk ´ pk crosses (1406, 1409, 1550). Spindle orientation is abnormal in the swollen asci, and no apical pore is formed (1625, 1679). Most mutant alleles are recessive for the ascus effect, but some are dominant with variable penetrance. Sorbose-resistant mutants at various loci act as dominance modifiers of the ascus effect of dominant alleles (1757). Sexual-phase-recessive allele C-1610 and dominant allele 17-088 are both associated with reciprocal translocations (1578).
pl : plug
VR. Linked to gran (0/75). Between asn (1%; 9%) and his-6 (16%) (1585, 1592, 2014).
Dense hyphae fill a 10-mm-diameter tube above the agar slant (1548). Because complex complete medium stimulates conidiation; scoring of morphology is clearer on minimal medium. The morphology is distinct from that of the closely linked mutant gran (1582).
plc-1 : phospholipase C-1
Unmapped
Cloned and partially sequenced: EMBL/GenBank U65686.
Encodes a PLC-like protein (phosphoinisotide-specific phospholipase, EC 3.1.-.-). Isogenes have been identified from short PCR fragments (EMBL/Genbank U65687 and U65688). Mutants of plc-1 obtained by RIP are highly sensitive to chemical agents that block polymerization of b-tubulin. Also sensitive to H2O2, which suggests that plc-1 is related to a transduction pathway stimulated by oxidative stress (1037).
pma-1 : plasma membrane ATPase-1
IIR. Between cya-4 (2T/18 asci) and preg (1T/15 asci (1447). Initially assigned incorrectly to linkage group I (813) as a result of the weak cross-hybridization shown with eat-2 (1925).
Cloned and sequenced: Swissprot PMA1_NEUCR, EMBL/GenBank J02602, M14085, PIR A26497, PXNCP, GenBank NEUATPPM, NEUATPASE; EST NM8D11.
Structural gene for plasma membrane ATPase (proton pump) (EC 3.6.1.35) (3, 813, 1427, 1922). Extensive electrophysiological studies defining the role of the pump (762, 763, 1921, 1923, 1924, 2189). Extensive biochemical studies: residues important for catalysis (1033, 1384, 2027); ligand binding at the active site (179); role in potassium ion symport (186, 188); N-ethylmaleimide sensitivity (352, 444, 1534). Conformational changes arising from ligand binding (759, 1257). Reconstitution in vesicles (1607) and stoichiometry of proton translocation (1608). Reconstituted ATPase is functional as a single-subunit monomer (758, 1804); functional size determination by radiation inactivation (207). Comparison with mitochondrial and vacuolar proton pumps (211). Secondary-structure analysis (868, 2158) and structural determination by electron crystallography (73, 437, 1106, 2004). Studies of biosynthesis, integration into the plasma membrane and enzyme topography (1, 4-6, 869, 1195-1197, 1258, 1697, 1698, 1803, 1805, 2159). Expression and mutagenesis studies in Saccharomyces cerevisiae (1248, 1249). Strong resemblance to a homolog in the pathogen Histoplasma capsulatum (1808). Mutants have been obtained that are inhibited by concanamycin A at high pH (213)
pmb : permease basic amino acid
IVR. Between uvs-2 (8%), T(S4342)R and Tip-IVR (513, 1578, 1776).
Cloned: Orbach-Sachs clone X7E5 (826).
Defective in basic L-amino acid transport system III as defined in ref. (1522); uptake of L-arginine, L-lysine, and L-histidine is reduced (1522, 2090, 2233) (Fig. 47). Used extensively for transport studies; see ref. (2231). Altered surface glycoprotein (2016). Selected as resistant to L-canavanine (1736, 2233). Scored on 1 mg/ml thiolysine or by resistance to a-difluoromethylornithine (464). Allelic with bat (1776), which was selected in arg-12s; pyr-3 (CPS-ACT+) by its ability to grow on minimal medium plus arginine when the parental double mutant was not able to grow because of arginine uptake and feedback onto the arginine-specific CPSase (2088). Probably allelic with bm-1 (linked to pyr-2, 24%), which was selected by canavanine resistance (1782). Possibly allelic with basa, which was selected by the inability of his-3 to grow on histidine plus methionine (1243). A mutant called argR, which maps right of pyr-2 (14%) and is probably allelic with pmb, makes lys-1 resistant to inhibition by L-arginine in the double mutant lys-1; argR (1082, 1083). Called Cr-10, Pm-B, pm b, UM-535, can-37.
pmg : permease general amino acid
IIL. Linked to pyr-4 (0/207). Right of ro-3 (4%) (29%)(1574).
Cloned and sequenced: Orbach-Sachs clone G3D3 (204).
Specifies the general amino acid permease, homologous to GAP1 of Saccharomyces (204). The mutant is greatly reduced in general amino acid transport [system II as defined in ref. (1521)] (Fig. 47). Uptake of arginine and phenylalanine is reduced. Selected by resistance to p-fluorophenylalanine in a neutral (system I), basic (system III) double mutant on medium lacking ammonium ions, where system II would be derepressed in wild type (513, 1693, 1694). A nonmetabolizable substrate has been found that is specific for this transport system (1488).
pmn : permease neutral amino acid
Allelic with mtr. (Also called Pm-N.)
Pogo (Transposable Element)
VR. Adjoins Tel-VR. Imperfect copies of the Pogo sequence are also present at nonterminal positions elsewhere in the genome and in different locations in different genetic backgrounds (1809, 1811).
Cloned and sequenced: EMBL/GenBank M37064, GenBank NEUTELVRA.
A retrotransposon-type element discovered as a 1.6-kb sequence adjoining the VR telomere. Contains direct terminal repeats and an open reading frame that is similar in sequence to reverse transcriptases (1809, 1811).
poi-1 : plenty of it-1
VR. Linked to cyh-2 (0T/18 asci) (1452).
Cloned and partially sequenced: EMBL/GenBank AA897897, AA898529; EST NP2A8.
The most abundant cDNA in starved mycelial and perithecial tissues. A novel sequence (1052).
poi-2 : plenty of it-2
VIIR. Linked to cat-2 (0 or 1T/18 asci), cox-8 (1T/18 asci) (1452).
Cloned and partially sequenced: EMBL/GenBank AA897974, AA898152; EST NM2H2.
The second most abundant cDNA isolated from starved mycelial and perithecial tissues. A novel sequence (1052).
polD : DNA polymerase delta
VR. Between ure-2 (<70kb) and am (10kb) (222).
Cloned and sequenced: Orbach-Sachs clone G9A10.
Encodes a sequence homologous to Drosophila pold and to DNA polymerase d from other species (222).
por : porin
Unmapped.
Cloned and sequenced: Swissprot PORI_NEUCR, EMBL/GenBank X05824, GenBank NCPORIN, PIR S07195, MNMCP; EST NC1D11.
Porin, also called VDAC (voltage-dependent, anion-selective channel), is the major protein of the mitochondrial outer membrane (1071). Most metabolites enter the mitochondrion through the channel formed by this polypeptide (1260, 1261, 1644). The porin polypeptide is unusual because it does not require an electrochemical potential across the inner mitochondrial membrane for import (675). Also, unlike signals of most polypeptides that are targeted to mitochondria, the import signal of porin appears to be at the C-terminus, not the N-terminus (416).
pp : protoperithecia
Unmapped.
Does not form protoperithecia. Impairs ascospore germination. Enhances growth of gpi on glucose or sucrose (1401). Obtained as a double mutant with gpi. No information is available on linkage or allelism with already mapped loci that affect ascospore viability or the formation of protoperithecia (e.g., ff, fs, gul-3, -4, -5, le-1, -2).
pph-1 : protein phosphatase-1
IV. Linked to mtr (0/18), arg-14 (1/18) (1447, 2271).
Cloned and sequenced: Swissprot P2A1_NEUCR, EMBL/GenBank X83593, NCPPH1, PIR S60471.
Structural gene for serine-threonine protein phosphatase PP2A catalytic subunit (EC 3.1.3.16) (2271). Essential function; the null mutant obtained by RIP can be harbored in a heterokaryon (2271). Level of mRNA is highest during conidial germination (2271). Characterization of activity (2297). Reduced expression is associated with reduced hyphal growth (2270).
pph-2 : protein phosphatase-2
Allelic with cna-1.
pph-3 : protein phosphatase-3
Unmapped.
Cloned and sequenced: Genbank AF049853 (2298).
Encodes serine-threonine protein phosphatase type 1 (PP1) (2298).
ppt-1 : phosphoprotein phosphatase-1
VR. Linked to inl (0/18) (1447).
Cloned and sequenced: EMBL/GenBank U89985.
Encodes a protein phosphatase of the PPT/PP5 family. Transcript is abundant in dormant conidia; transcript level declines during germination (2272). For a review of the protein phosphatases identified in filamentous fungi, see ref. (535).
ppz-1 : protein phosphatase Z
Changed to pzl-1.
pr(Sk-2) : partial resistance to Sk-2 killing
IIIR. Between ser-1 and ad-4. Within the recombination block that is associated with Sk-2 (2121).
Confers partial resistance to the killing of ascospores by Sk-2 (2121).
prd : period
Mutations at numerous loci other than frq have been shown to affect period length of the circadian clock. Those named period were identified on this basis. Period length and/or temperature compensation are also affected by mutations at loci initially recognized on the basis of quite different characteristics, e.g., cel, chol-1, fas, phe-1. Reviewed in refs. (560), (1130), and (1133).
prd-1 : period-1
III. Linked to acr-2 (5%), Cen-III (0T/35 asci), pro-1 (20%) (619, 710).
Altered period of circadian conidiation rhythm. One allele is known, with a 25.8-hr period at 25ºC without csp (619, 711). Recessive. Grows at 60% of the wild-type rate. Period lengths of double mutants with frq-1, -2, -3 (619). Temperature compensation (713). Altered membrane fatty acid composition (413); effects of fatty acid addition to the cel mutant are not observed in the cel; prd-1 double mutant (1132). Membrane cytochromes appear to be similar to wild type (198). Name changed from frq-5 (711). For reviews of circadian mutants, see the entry for frq.
prd-2 : period-2
VR. Between lys-2 and am (713).
Cloned (1184).
Altered period of circadian rhythm (25.5 hr at 25ºC without csp, for allele IV-2). Slower than normal growth. Recessive (621). Temperature compensation described (713). Synergistic interactions with frq long-period alleles [discussed in ref. (1382)]. Called IV-2.
prd-3 : period-3
I. Near Cen-I (713).
The period of circadian rhythm is altered (25.1 hr at 25ºC without csp, for allele IV-4) (621). Slower than normal growth. Recessive. Temperature compensation described (713). Called IV-4.
prd-4 : period-4
IR. Between os-1 (2%) and arg-13 (3%) (1212).
Cloned: pSV50 clone 9:2H.
The period of circadian rhythm is altered (18.0 hr at 25ºC without csp, for allele V-7) (621). Normal growth rate. Dominant, based on heterokaryons with different ratios of wild-type and mutant nuclei (56). Temperature compensation described (713). Called V-7.
prd-5 : period-5
IIR. Between arg-12 (4%; 10%) and un-20 (8%). Linked to alc-1 (8%) (1186).
Altered period of circadian conidiation rhythm. One allele is known, with a 19.5-hr period at 25ºC. Whereas temperature compensation is normal, expression of the mutant phenotype is temperature- dependent. The mutation is pleiotropic, affecting not only the circadian period but also morphology, sexual development, linear growth rate, tolerance to elevated temperature, and acriflavin resistance. Possibly allelic with ff-1 (1186).
prd-6 : period-6
VR. Linked to ad-7. Between inl (9%; 30%) and pab-2 (2%), inv (£1%), asn (7%) (1382).
The circadian period length is shortened and is temperature-sensitive. Epistatic to prd-2 (1382).
preg : phosphatase regulation
IIR. Between T(NM177)L, nuc-2 (pcon) (1% or 2%), and arg-12, T(NM177)R (1157, 1333). Linked to cit-1 (0T/18 asci), nuo78 (0T/17 asci) (1447).
Cloned and sequenced: Swissprot PREG_NEUCR, EMBL/GenBank L07314, PIR S52974, EMBL NCPREGPRO, GenBank NEUPREGPRO; pSV50 clones 2:8E, 2:9F.
Encodes a cyclin. Regulator of repressible alkaline phosphatase and other steps in phosphorus uptake and metabolism. Hypostatic to nuc-1, epistatic to nuc-2. The pregC constitutive mutant is recessive to its wild-type allele (1322, 1325, 1333). Scored on high phosphate medium by a staining reaction with α-naphthyl phosphate plus Diazo Blue B (747, 2096). Used to study phosphate transport (1229). For regulation model, see refs. (1325), (1326), and (1331) and Fig. 51.
Prf : Perforated
VR. Linked to al-3 (3%), probably to the right (1678).
Many of the asci in crosses heterozygous for Prf produce asci with 8-12 small apical pores and variable numbers of large, multinucleate ascospores. A majority of asci cut out a single giant ascospore (Fig. 52). A few asci develop normally and produce small ascospores that can be used for mapping. Prf is recessive- lethal in the vegetative phase and must be maintained in a heterokaryon. The first-formed croziers appear normal and differentiate asci, but croziers that are formed later revert to mitosis and become multinucleate, with synchronous nuclear divisions similar to those in Ban (1678).
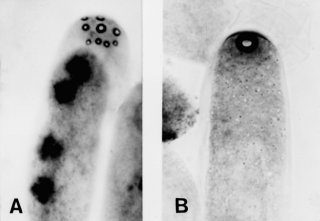
FIGURE 52 (A) The tip of an ascus from a cross heterozygous for the mutant gene Prf: Perforated, showing multiple apical pores. (B) A normal ascus, showing the single apical pore that is characteristic of wild type. Reprinted from ref. (1678 ), with permission from Mycologia, Vol. 79, p. 702. Copyright 1987 The New York Botanical Garden.
prl-1 : mitochondrial proteolipid ATP synthase
Allelic with oli-1.
pro : proline
Three pro loci are defined by mutant genes. pro-3 and pro-4 are putative isogenes for the first step in proline biosynthesis, the conversion of glutamate to glutamate semialdehyde. The next step, catalyzed by g-glutamyl phosphate reductase (GPR) (EC 1.2.1.41) (glutamate-5-semialdehyde dehydrogenase) (glutamyl-g-semialdehyde), is unrepresented by a mutant or a sequence. The final step (EC 1.5.1.2) is defined by pro-1. As part of the greater arginine biosynthetic pathway (Fig. 10), pro-3 and pro-4 originally were called arg-8 and arg-9.
pro-1 : proline-1
IIIR. Between sc (7%), ser-1 (1%; 114 kb; 3T/38 asci) and ace-2 (2%; 36 kb) (449, 940, 1122, 1587).
Cloned and sequenced: EMBL/GenBank U30317, PIR S57863; pSV50 clones 13:10A, 18:2C, 21:6D (449)
Structural gene for pyrroline-5-carboxylate reductase (EC 1.5.1.2) (449, 2291). Uses proline but not ornithine, citrulline, or arginine (1967) (Fig. 10). Transcription of pro-1 appears to be independent of cpc-1-mediated cross-pathway control (449).
pro-3 : proline-3
VR. Linked to inl, pab-1 (0/74). Between his-1 (4%) and pk (2%; 6%) (1585).
Uses proline, ornithine, citrulline, or arginine (1964). Blocked in the reduction of glutamic acid to glutamic semialdehyde (2165), which is catalyzed by glutamate 5-kinase (EC 2.7.2.11) (g-glutamyl kinase) (Fig. 10). Arginine and citrulline are used via the arginine catabolic pathway (arginase and ornithine transaminase), ornithine via ornithine transaminase (310, 453, 2164, 2165). Tends to accumulate second mutations, including arg-2, his-1 (116, 1932). Ability to grow on arginine is modified by ipa (1932) and ota (453). Suppressed by arg-6 mutations that affect N-acetylglutamate kinase (383, 2204). Called arg-8.
pro-4 : proline-4
IIIR. Linked to thi-2 (0/78), ota (4%) (108, 1603).
The proline pathway is blocked in the reduction of glutamic acid to glutamic semialdehyde (2165), glutamate 5-kinase (EC 2.7.2.11) (g-glutamyl kinase) (Figs. 10 and 11. Uses proline, ornithine, citrulline, or arginine (1967). Citrulline and arginine are used via the arginine catabolic pathway (arginase and ornithine transaminase) and ornithine via ornithine transaminase (310, 2164, 2165). Leaky. Called arg-9.
prol : proline
Symbol changed to pro.
Protein Kinases
Protein phosphorylation is required for fundamental functions such as the cell cycle, transcription, and mating. For a review of protein kinases in filamentous fungi, see ref. (535). For genes that specify protein kinases in Neurospora, see cna, cot-1, mad, mak, mcb, mdk, mek, mgk, mik, msk, nik, nrc, pph-1, pzl-1, and rgb-1. Numerous ESTs have been identified that appear to identify protein kinases.
Protein Phosphatases
The protein phosphatases identified in filamentous fungi are reviewed in ref. (535). Genes specifying protein phosphatases in Neurospora include cna-1, cyt-4, pph-1, ppt-1, pzl-1, and rgb-1.
prt : protease
Allelic with pts.
ps15-1 : phosphorylation of PS15 protein
Symbol changed to psp.
psi-1 : protein synthesis initiation-1
IVR. Between Cen-IV and T(ALS159), pyr-1 (4%) (1220, 1546, 1578).
Conidial germination and hyphal growth are inhibited at 35ºC but are normal (or nearly so) at 20ºC. Protein synthesis is reduced after a shift to restrictive temperature. Recessive in heterokaryons (1220). Scored as an irreparable heat-sensitive un mutant (see un). Germinated ascospores die after 3 days at 34ºC. Used to examine the effects of inhibiting protein synthesis on heat shock (2047).
psp : phosphorylation of small protein
Allelic with ndk-1 (1489a).
pt : phenylalanine + tyrosine
IVR. Between pdx-1 (2%), T(S1229)L and col-4 (2%) (87, 111, 1580).
Evidently the structural gene for chorismate mutase (EC 5.4.99.5) (87, 601) (Fig. 13). Allele NS1 has thermolabile chorismate mutase (317). Requires phenylalanine plus tyrosine (396). Inhibited by complex complete medium. NS1strains are temperature-sensitive, growing on minimal medium at 25ºC, where they are readily scorable by blue fluorescence under long-wave UV and by browning of medium in aging cultures (2013). The original strain, S4342, contained the linked but separable insertional translocation T(S4342), the presence of which should not change conclusions regarding gene order in ref. (87).
pts-1 : protease-1
Unmapped. Segregates as a single gene not closely linked to alcoy markers (40 isolates) (831).
Structural gene for carbon-, nitrogen-, and sulfur-controlled extracellular alkaline protease (EC 3.4.21.-). The allele found in strain Groveland-1c a (FGSC 1945) encodes a fast electrophoretic variant that is synthesized under conditions of limiting C, N, and S. Regulation reviewed refs. (1281) and (1325). Called prt (831).
Punt (Transposable element)
IVR. A RIP-modified copy (PuntRIP1) is present in the Fsr-63 pseudogene of Oak Ridge strains, between Cen-IV and pyr-1. Unmapped copies are also present at other locations. One of these, dPunt, is a defective copy that has apparently not been modified by RIP (1266).
Cloned and sequenced: EMBL/GenBank AF181821 (PuntRIP1); AF181822 (dPunt).
An inactive element bounded by imperfect terminal inverted repeats and a 3-bp target-site duplication. Most closely allied to the Fot1 family of elements thought to transpose through a DNA intermediate (439). Discovered as a 1.9-kb insertion in Fsr-63. Similar sequences are present in other Neurospora crassa strains and in Neurospora sitophila (1266).
Purine
See ad, gua.
put-1 : putrescine-1
Symbol changed to spe-1.
puu-1 : putrescine uptake-1
IVR. Between arg-2 (5%) and cot-1 (8%) (472)
Cloned and sequenced: Orbach-Sachs clone G17G02.
Calcium modulation of polyamine transport is lost (472). The mutant is temperature-sensitive (37ºC) and osmotic remedial (1 M sorbitol) (451). Used to study polyamine toxicity (469, 478).
pyr : pyrimidine
All pyrimidine auxotrophs in Neurospora are nonspecific, responding to any pyrimidine nucleoside, nucleotide, or base. The symbol pyr therefore is used for genes concerned with the biosynthetic pathway. Nucleosides or nucleotides are more effective than corresponding bases as growth supplements for pyr-1 (1223) and apparently for other pyr mutants. However, after a lag uracil is used nearly as effectively as uridine (1363). No cytidine- or thymidine-specific requirement exists, because Neurospora lacks thymidine kinase (792) and because any exogenous pyrimidine supplement is cycled back through uridine monophosphate, which provides all normal end products of pyrimidine biosynthesis (2217). For this reason, DNA cannot be specifically labeled by supplying [3H]thymidine under normal circumstances [but see tk entry and ref. (1772)]. Mutants have been obtained (uc-2, -3, -4, and -5, ud-1) that block the pathway back through UMP and so prevent general labeling from a single precursor (2217). Cytosine in DNA can be labeled specifically by the method of ref. (2246). For a general review of pyrimidine metabolism, see ref. (1487). For systematic gene-enzyme work, see refs. (288) and (291). For the pyrimidine biosynthetic pathway, see Fig. 53. For loci concerned with pyrimidine salvage or pyrimidine uptake, see Fig. 65. Complex interactions between lys and pyr mutants have been described (910).
Regulation: Pyrimidine biosynthetic enzymes differ in their mode of regulation. The CPS(pyr)-ACT complex is feedback-inhibited by UTP and derepressed by end-product depletion, but is insensitive to repression in the fluoropyrimidine-resistant mutant fdu-2 (267, 289). Dihydro-orotase is relatively unresponsive to end-product limitation, and dihydro-orotate dehydrogenase is induced by a precursor that is probably, by analogy with yeast, dihydro-orotate, the substrate of the enzyme. Regulation of the last two enzymes has not been studied systematically. Pyrimidine regulation of the uptake and salvage pathways of pyrimidine is discussed under individual loci; see uc-5, ud-1. Many aspects of pyrimidine metabolism are under the control of general nitrogen metabolite regulation (266) (Fig. 50).
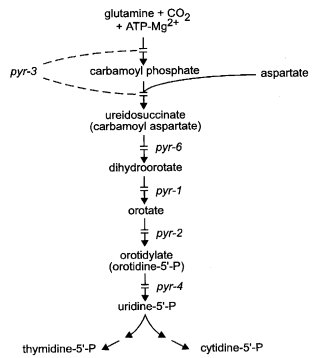
FIGURE 53 The pyrimidine biosynthetic pathway, showing sites of gene action (288 , 875 , 1655 , 2216 ). Carbamoyl phosphate (CAP) for arginine synthesis is made as a separate pool by a different enzyme system (see arg-2, arg-3 in Fig. 10). Interchange between the two pools occurs only in certain mutant combinations. From ref. (1596 ), with permission from the American Society for Microbiology.
pyr-1 : pyrimidine-1
IVR. Between psi (4%), T(ALS159) and pdx-1 (<1%; 10%) (1369, 1580, 1582).
Requires uracil or another pyrimidine. Lacks dihydro-orotate dehydrogenase activity (EC 1.3.99.11) (288, 291) (Fig. 53). Ascospores from pyr-1 ´ pyr-1 are black if the crossing medium is supplemented with 1 mg/ml uracil, but white and inviable with 0.1 mg/ml (1254).
pyr-2 : pyrimidine-2
IVR. Between nit-3 (2%; 9%) and rug (3%), T(NM152R (1255, 1582, 1585, 1950).
Requires uracil or other pyrimidine. Lacks orotidine 5'-monophosphate pyrophosphorylase activity (EC 2.4.2.10) (288, 291) (Fig. 53). Needs 0.5 mg/ml uracil in medium for optimal growth. Allele 38502 is leaky.
pyr-3 : pyrimidine-3
IVR. Between T(S1229)R arg-14, cyt-19 (5%), T(NM152)L and his-5 (1%) (457, 1377, 1580).
Requires uracil or other pyrimidine (1363). Growth is inhibited by purine nucleosides and nucleotides (1621). Structural gene for pyrimidine-specific carbamyl phosphate synthase (CPS, EC 6.3.5.5) and aspartate carbamyl transferase (ACT, also abbreviated ATC, EC 2.1.3.2) (875, 1664) (Fig. 53). Mutants may lack either or both activities, e.g., alleles KS43 (CPS+ACT-), KS20 (CPS-ACT+), and KS11 (CPS-ACT-) (2216). Unlike yeast, no feedback-insensitive CPS+ACT+ mutants have been discovered in Neurospora (1659). Some mutants have kinetically altered ACTase (875, 1720). Used extensively for studies of channeling and of the relation of gene structure to the two enzyme activities (455). Normally, carbamyl phosphate (CAP) produced by pyr-3+ is used solely for pyrimidine synthesis, and CAP produced by arg-2+ and arg-3+ is used for arginine synthesis, the enzymes being in different organelles; however, a deficiency of the next enzyme in either pathway permits the overflow of CAP into the other pathway [reviewed in ref. (455)]. Hence, CPS-ACT+ alleles are suppressed by arg-12s (476), and CPS+ACT- alleles can be selected as suppressors of arg-2 and arg-3 (1311, 1734). Some of the CPS+ACT- alleles, called pyrsu-arg, suppress the arginine requirement but retain enough ACTase activity that they have no detectable pyrimidine requirement (1717, 1722). arg-13, arg-4, -5, -6, and am partly suppress CPS-ATC+ alleles (1313). The fertility of interallelic crosses is variable and often very poor (1311). Complementation is good between CPS-ACT+ and CPS+ACT- mutants (476) and between some pairs of CPS+ACT- mutants; otherwise, complementation is poor (1665, 2241). Complementation maps (1311, 1665, 1717, 2241). Fine-structure map (1666, 2030). Mutational analysis (1667). The direction of translation, based on enzyme types of polar mutants, is from CPS to ACT (1664). Allele 37815(t) is heat-sensitive (34 vs 25ºC) (136). Allele 1298 is CO2-remediable (363, 364). Strain KS12, a pyr-1 pyr-3 double mutant, originally was called pyr-5 (648). The different classes of pyr-3 alleles have been called M (CPS-P-less), N (ACT-less), and MN (lacking both activities) (Figs. 11 and 53).
pyr-4 : pyrimidine-4
IIL. Between het-c (1%), Pad-1 (1%) and ro-3 (1% or 2%) (1109, 1414, 1592)).
Cloned and sequenced: Swissprot DCOP_NEUCR, EMBL/GenBank M13448, X05993, GenBank NCPYR4G, NEUPYR4, PIR A24398, DCNCOP.
Structural gene for orotidine 5'-monophosphate decarboxylase (EC 4.1.1.23) (288, 291, 1655) (Fig. 53). Requires uracil or other pyrimidine. Fertile crosses homozygous for pyr-4 can be made using very high levels of uridine (15-20 mg/ml) (1424).
pyr-5 : pyrimidine-5
Used for a pyr-1 pyr-3 double-mutant strain (648).
pyr-6 : pyrimidine-6
VR. Between asn (6%) and un-9 (2%) (321, 1603).
Requires uracil or other pyrimidine. Lacks dihydro-orotase activity (EC 3.5.2.3) (288, 291) (Fig. 53). When given a small amount of uridine, a strain carrying the only allele (DFC37) pauses and then grows well beyond the level normally supported by the supplement; at no time is dihydro-orotase detectable (288).
pyrG : pyrimidine G
From Aspergillus nidulans.
Introduced into Neurospora in plasmid ppyrG and used as a selectable marker for transformation (2131). Both pyrG of Aspergillus and pyr-4 of Neurospora encode orotidine-5'-phosphate carboxylase. Each gene functions in the heterologous species. The Aspergillus gene is not subject to RIP when single-copy transformants of Neurospora are used in crosses.
Pyruvate Decarboxylase
See cfp.
pzl-1 : phosphatase-Z-like-1
IL. Linked to mus-40 (0/18), cyt-21 (1/18), nit-2 (3/16) (1447).
Cloned and sequenced: EMBL/GenBank AF071751, AF071752.
Encodes a novel type-Z-like Ser/Thr protein phosphatase expressed in all vegetative stages of development (2035). Called ppz-1. For a review of protein phosphatases in filamentous fungi, see ref. (535).
Return to the 2000 Neurospora compendium main page