q : quinolinic acid
Symbol changed to nic-1.
qa Cluster
VIIL. Between ars-1, tRNALEU and Cen-VII, met-7 (0.2%; 1%) (304, 340, 1725).
Cloned and sequenced: EMBL/GenBank X14603, GenBank NCQA. Included in VII contigs GenBank AC006498.
The cluster consists of seven genes concerned with the uptake and catabolism of quinate and with regulation of genes in this pathway (730, 734) (Figs. 54 and 55). The qa-1F and qa-1S genes are regulatory. The order on the conventional genetic map is: tRNALEU qa-1F qa-1S qa-Y qa-3 qa-4 qa-2 qa-X Cen-VII met-7.

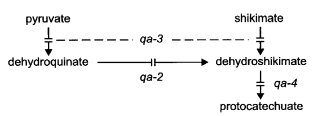
FIGURE 54 The quinate (aromatic amino acid) catabolic pathway, showing sites of gene action (304 , 306 , 728 , 795 , 983 ). qa-1 is a regulatory gene affecting all three structural genes. qa-2+ activity (catabolic dehydroquinase) can be replaced by the product of aro-9+, the equivalent gene in the aromatic biosynthetic pathway. See ref. (795 ) for the catabolic steps subsequent to protocatechuic acid. From ref. (1596 ), with permission from the American Society for Microbiology.
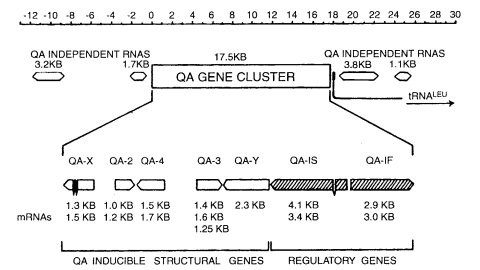
FIGURE 55 A transcriptional map of the 42-kb region containing the qa gene cluster and adjacent regions. The 17.3-kb region of the qa gene cluster is defined by the seven quinic acid-inducible mRNA transcripts shown. Other RNAs, including tRNALEU, show no induction and therefore are not part of the cluster. The direction of transcription for each qa gene is indicated. The sizes of discrete mRNA transcripts on Northern blots detected by DNA-RNA blot hybridization are given immediately below each gene. The positions of short introns in qa-X and qa-1S are indicated. The order is reversed in this drawing relative to that of loci on the conventional genetic map, where tRNALEU adjoins qa-1F on the left and qa-1S adjoins Cen-VII on the right. Copied from ref. (730 ), with permission from the American Society for Microbiology.
qa-1F : quinate-1F
VIIL. Leftmost gene in the qa cluster, between tRNALEU and qa-1S (304, 723, 1725).
Cloned and sequenced: Swissprot QA1F_NEUCR, EMBL/GenBank X14603, GenBank NCQA, PIR F31277, S04256. Included in VII contigs GenBank AC006498.
Unable to use quinate or shikimate as the sole carbon source. Regulatory (345). Activator of the quinate catabolic pathway (723). Fast complementing. Effects of expression on chromatin structure (132). Characterization of binding sites in the qa cluster (131). Interaction with QA-1S; evolutionary origin (859) (Fig. 55).
qa-1S : quinate-1S
VIIL. Between qa-1F and qa-Y in the qa gene cluster (304, 723, 1725).
Cloned and sequenced: Swissprot QA1S_NEUCR, EMBL/GenBank X14603, GenBank NCQA, NCQA1SRA, PIR E31277, S04255. Included in VII contigs GenBank AC006498.
Unable to use quinate or shikimate as the sole carbon source. Regulatory (345). Repressor of the quinate pathway (723). Slow complementing. Effects of expression on chromatin structure (132). Effect of deletion on qa expression (298). Interaction with QA-1F; evolutionary origin (859) (Fig. 55).
qa-2 : quinate-2
VIIL. Between qa-4 and qa-X in the qa gene cluster (304, 723, 1725).
Cloned and sequenced: Swissprot 3DHQ_NEUCR, EMBL/GenBank X14603, GenBank NCQA, NCQAX2, PIR A31277, S04251. Included in VII contigs, GenBank AC006498.
Structural gene for catabolic dehydroquinase (EC 4.2.1.10) (38, 306, 723, 983, 1725). The double mutant aro-9; qa-2 is unable to use quinate or shikimate as the sole carbon source (1726). [aro-9 lacks biosynthetic dehydroquinase. The single mutant aro-9; qa-2+ grows on minimal medium without supplement (Figs. 54 and 55).] qa-2 is scored conveniently as an aro auxotroph when aro-9 is present. The qa-2+ gene cloned in pBR322 is expressed constitutively from its own promoter in Escherichia coli (39, 729, 857). Used as a selectable marker for Neurospora crassa transformation (341) and as a reporter (435). The qa-2 promoter has been used to regulate the expression of recombinant genes (57, 2074). Allele M246 is stable (308). Homozygous fertile with black ascospores. Position-dependent effects on gene expression (2148). Effects of rearrangements in the promoter region on activation (724). N. crassa and Neurospora africana differ in the intergenic region between qa-X and qa-2; the differences have been examined for their effects on qa-2 expression in N. crassa (65).
qa-3 : quinate-3
VIIL. Between qa-Y and qa-4 in the qa gene cluster (304, 343, 723).
Cloned and sequenced: Swissprot DHQA_NEUCR, EMBL/GenBank X14603, GenBank NCQAX3, PIR S04253, C31277. Included in VII contigs GenBank AC006498.
Unable to use quinate or shikimate as the sole carbon source (344). Structural gene for quinate (shikimate) dehydrogenase (EC 1.1.1.24) (344). Revertants have altered enzymes. Transcription is in the direction qa-4 to qa-1 (307) (Figs. 54 and 55).
qa-4 : quinate-4
VIIL. Between qa-3 and qa-2 in the qa gene cluster (304, 343, 723).
Cloned and sequenced: Swissprot 3SHD_NEUCR, EMBL/GenBank M10139, X14603, GenBank NCQA4, PIR A22421, D31277, S04252. Included in VII contigs GenBank AC006498.
Unable to use quinate or shikimate as the sole carbon source (344). Structural gene for dehydroshikimate dehydrase (EC 4.2.1.-) (344, 723, 1761) (Figs. 54 and 55).
qa-X : quinate-X
VIIL. Between qa-2 and met-7 in the qa gene cluster (723).
Cloned and sequenced: Swissprot QAX_NEUCR, EMBL/GenBank X14603, PIR B31277, S04250, GenBank NCQA. Included in VII contigs GenBank AC006498.
Part of the quinate complex. Function unknown. Figure 55.
qa-Y : quinate-Y
VIIL. Between qa-1S and qa-3 in the qa gene cluster (723).
Cloned and sequenced: Swissprot QAY_NEUCR, EMBL/GenBank X14603, PIR G31277, S04254, GenBank NCQA. Included in VII contigs GenBank AC006498.
Encodes quinate permease (Fig. 55). Growth characteristics of deletion mutant (298).
qcr7 : ubiquinol-cytochrome c reductase 7
Unmapped.
Cloned and sequenced: Swissprot UCRQ_NEUCR, EMBL/GenBank U20790; EST SC1H8.
Encodes subunit VII of ubiquinol-cytochrome c oxidoreductase (EC 1.10.2.2), an 11-kDa ubiquinone-binding protein homologous to Saccharomyces QCR8 and mammalian subunit VII. The eight N-terminal residues of a precursor protein are cleaved to produce a mature product; these residues appear to be important for function but not for directing the polypeptide to the mitochondrion (1217).
qcr8 : ubiquinol-cytochrome c reductase 8
Unmapped.
Cloned and partially sequenced: EMBL/GenBank AI399120; EST W10H7 (1452).
Specifies ubiquinol-cytochrome c oxidoreductase complex III subunit VIII (EC 1.10.2.2), an 11-kDa ubiquinone-binding protein.
qde : quelling defective
Defective in the transgene-silencing mechanism known as “quelling” (392). Quelling reduces vegetative expression when multiple copies of a gene are introduced into the genome by transformation (390, 392, 394, 1737). Quelling occurs posttranscriptionally and appears to affect mRNA stability. For a critique relating quelling to silencing in other organisms, see ref. (130).
qde-1 : quelling defective-1
Unmapped.
Cloned and partially sequenced: EMBL/GenBank AJ133528.
Specifies a product resembling RNA-dependent RNA polymerase in the tomato (391). Suppresses quelling of al-1 duplications (392).
qde-2 : quelling defective-2
Unmapped.
Cloned and sequenced (310a).
Specifies a product homologous to RDE-1 of Caenorhabditis, which is essential for gene silencing by double-stranded RNA in that organism (310a). Suppresses quelling of al-1 duplications (392).
qde-3 : quelling defective-3
Unmapped.
Cloned and sequenced: GenBank AF205407 (393a).
Specifies a RecQ DNA helicase (393a). Suppresses quelling of al-1 duplications (392).
qde-4 : quelling defective-4
Unmapped.
Suppresses quelling of al-1 duplications (393).
Return to the 2000 Neurospora compendium main page