R : Round spore
IR. Between aro-8 (4%) and T(MD2)L, het-5, un-18 (11%) (1578, 2129).
All eight ascospores of heterozygous R/R+ asci are round rather than ellipsoid (Fig. 56). R is thus nonautonomous (or nearly so) in ascospores and dominant in the ascus (1367). Ascospores are round even in nonlinear asci (1546, 1966). A single germination pore is formed in spores that are completely round, two pores in spores that are slightly ovoid (1483a). Vegetative morphology is abnormal, somewhat resembling that of peach. Initial growth on a slant is concentrated around the inoculation point. The vegetative morphology is recessive in heterozygous duplications, such as those from T(NM103) (2123). Female sterile with no perithecia, but R ´ R crosses are productive if R is heterokaryotic in the female parent (1962). Used to study duplication instability (2123) and to show that ascospore development is autonomous in individual asci of different genotype within the same perithecium (999). Used to show that defective asci in crosses between inbred strains do not result from self-fertilization of mat A ´ mat A or mat a ´ mat a (1687). Recurrences have been obtained (1962, 1966). Although the R locus is not included in duplications from T(MD2) ´ Normal, round ascospores that appear to carry new R mutations are found among the progeny of crosses in which one parent is the breakdown product of such a duplication (978). For other genetically determined round ascospores, see ref. (117). Called Rsp (1966), but the original symbol R has priority. [The symbol rsp has been used for cytoplasmically determined respiration-deficient mutants (1741)].

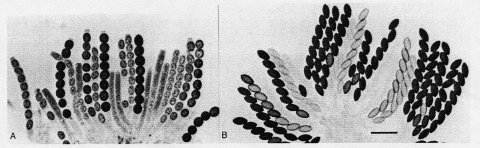
FIGURE 56 (A) Asci from a cross of wild type X Round spore. The Round spore mutation is ascus-dominant: all eight ascospores (R as well as R+) are round. (B) Asci from a cross between two wild-type strains, for comparison. The asci with unpigmented or lightly pigmented spores are immature. Bar length = 50 mm. Photographs by N. B. Raju. From refs. (1676 ) and (1680 ), with permission from Urban & Fischer Verlag and from the British Mycological Society, respectively.
r(Sk-2) : resistant to Sk-2
IIIL. Between cum (1% or 2%) and T(NM183) (2%), acr-7 (7%), sc (17%) (280, 2121, 2125, 2127).
Prevents killing by Sk-2 of ascospores that are not themselves Sk-2. Does not confer resistance to killing by Sk-3. Allelism of r(Sk-2) with Sk-2 is not excluded because recombination in this region is blocked when Sk-2 is heterozygous. Recombination is not blocked by r(Sk-2)-1. Allele P527 was found in Neurospora crassa from Louisiana. Only one other known N. crassa wild type is resistant (2121). For reference strains and tester strains for identifying the various killers and genes conferring resistance to killing, see refs. (2127) and (2128).
r(Sk-3) : resistant to Sk-3
IIIL. Between cum (4%) and acr-7 (17%) (280, 2121, 2125).
Prevents killing by Sk-3 of ascospores that are not themselves Sk-3. Does not confer resistance to killing by Sk-2. Allelism of r(Sk-3) with Sk-3 is not excluded because recombination in this region is blocked when Sk-3 is heterozygous. Recombination is not blocked by r(Sk-3). Found in Neurospora intermedia and introgressed into Neurospora crassa (2121). For reference strains and tester strains for identifying the various killers and genes conferring resistance to killing, see refs. (2127) and (2128).
Radiation Sensitivity
See Mei-2, mei-3, mus, nuh-4, phr, upr-1, uvs. The symbol rad has not been used in Neurospora.
ran : ran-like
Unmapped.
Cloned and sequenced: EMBL/GenBank AJ000287, GenBank NCRANRNA.
Shows homology with a GTP-binding protein of the ran family (1341).
rap-1 : ribosome-associated protein-1
IIR. Near Cen-II. In same cDNA with vma-6 (~1200 bp) (1317).
Cloned and sequenced: Swissprot RSP4_NEUCR, EMBL/GenBank U36470.
Encodes a putative 40S ribosome-associated protein of unknown function (1317).
ras : ras-like
Genes belonging to the ras supergene family (named for an oncogene of the rat sarcoma virus) encode highly conserved G proteins that are important for signal transduction pathways regulating growth and differentiation. The Neurospora ras-like genes called ras, krev, and ypt have been identified as ras-like by DNA sequence homologies. When phenotypes are examined using null mutants obtained by RIP, ras-like genes may prove to be allelic with already known mutants (e.g., the gene originally designated ras-2 proved to be allelic with smco-7).
ras-1 : ras-like-1
IVR Between met-5 (1/18) and nit-3 (2/28) (968).
Cloned and sequenced: Swissprot RAS1_NEUCR, EMBL/GenBank U33746, X53533, GenBank NCRAS, PIR S12892; EST SM1B4 .
Mutant strains and strains overexpressing ras-1 are female-sterile, with defective aerial hyphae and few conidia (971, 2021). The RIP-inactivated mutant shows very slow growth (945). Called NC-ras.
ras-2 : ras-like-2
Allelic with smco-7.
ras-3 : ras-like-3
VIIR. Immediately adjacent to thi-6. Present in a cosmid adjacent to that carrying cfp. Linked near un-10 on the RFLP map, but not borne on the same plasmid (1267).
Cloned and sequenced: Orbach-Sachs clones X18A06, G15F10, G22A09.
Encodes novel protein loops. Transcribed convergently with thi-6 (1267).
rca-1 : regulator of conidiation in Aspergillus-1
VR. Linked to cyh-2 (0T/16 asci) (1447, 1902).
Cloned and sequenced: EMBL/GenBank AF006202.
The Myb-like functional homolog of flb1 in Aspergillus nidulans. The Neurospora gene complements the sporulation defect of the Aspergillus mutant, but a deletion mutant has no visible effect on growth or conidiation in Neurospora other than to make early hyphal growth counter-clockwise or straight, in contrast to the typically clockwise spiral growth of Neurospora wild type (see Spiral Growth) (1902).
rco-1 : regulator of conidiation-1
III. Linked to acr-2 (7%; 11%) (2256).
Cloned and sequenced: Swissprot RCO1_NEUCR, EMBL/GenBank U57061; Orbach-Sachs clones X1B01, X14D07, X14F01.
Encodes a product homologous with Saccharomyces cerevisiae TUP1 (2256). Selected as enabling higher basal expression of a con-10::hph fusion gene during vegetative growth. Mycelial growth rates are reduced and conidiation is abnormal. Female sterile. Four alleles have four different conidiation morphologies. Presumptive RIP alleles are defective in the separation of conidia. Ascospores containing the null allele show counterclockwise spiral growth when germinated, in contrast to wild type, which shows clockwise spiral growth (see Spiral Growth) (2256). Affects the expression of con-10 and ccg-1 (1154).
rco-3 : regulator of conidiation-3
Allelic with dgr-3 and hence, with sor-4. See sor-4.
rDNA : ribosomal DNA
VL. Located in the NOR, between Tip-VL and mus-18 (0T/17 asci; 20 kb) (1447).
Cloned and sequenced: 5.8S sequence EMBL/GenBank X02447, M10692, NCRRN58S, EMBL NCRGB, GenBank NEURGB; 17S sequence EMBL/GenBank X04971, GenBank NCRRNAS; 18S sequence (partial) EMBL/GenBank M11033, EMBL NCRRE, GenBank NEURRE; 25S sequence (partial) GenBank X01373; ITS sequence EMBL/GenBank M13906, EMBL NCRGITR, Genbank NEURGITR; 28S-like sequence (partial) EMBL/GenBank M38154, EMBL NCRRNA1; Orbach-Sachs clone X15E04.
Genes specifying 5.8S, 17S, and 25S ribosomal RNA (but not 5S) are located in the NOR in a series of ~150 tandem repeats, each ~9.3 kb long. The repeats are separated by nontranscribed spacers (423, 672). rDNA is a site of chromosome breakage in the breakdown of duplication strains derived from translocations that involve the NOR (261). See NOR.
rec : recombination
trans-acting genes that regulate meiotic recombination in specific regions and constitute a class of recombination control genes first described in Neurospora (339, 990) (Fig. 57). Initially detected by changed intragenic recombination (up to 25´), but crossing over between loci may also be affected (up to 40X). High recombination is recessive. Products of dominant alleles (called rec+) are thought to repress the initiation of recombination at specific recombinator loci, of which cog is a well-studied example. A given target locus or region appears to be affected specifically by alleles at only one rec locus. Polarity of intragenic recombination in a target locus may be changed by the presence of the controlling rec+ allele. Control of recombination is independent from the control of gene expression (311). Three rec loci have been identified and characterized. Ten are estimated to be present in the Neurospora crassa genome. Differences at rec loci are present in commonly used laboratory wild types (338). For reviews, see refs. (316), (335), and (645).
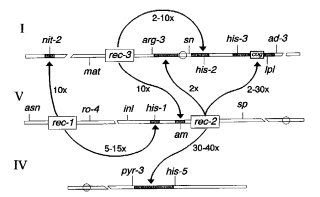
FIGURE 57 Linkage map showing the location and targets of action of rec-1, rec-2, and rec-3. Original figure from D. E. A. Catcheside.
rec-1 : recombination-l
VR. Between ro-4 (7%) and asn (5%) (325, 332).
Presence of allele rec-1+ reduces recombination within the loci his-1 (VR) (990, 2083) and nit-2 (IL) (320, 324) (Fig. 57), but does not affect recombination at other his loci (314). A recessive allele from another lineage was called rec-z until it was identified as rec-1 (324).
rec-2 : recombination-2
VR. Between sp (1%; 6%) and am (2%) (338, 1931).
Presence of dominant allele rec-2+ reduces recombination within the his-3 locus (IR) and reduces crossing over in the intervals pyr-3 to his-5 (IVR), his-3 to ad-3 (IR), and arg-3 to sn (IL) (336, 338, 1930) (Fig. 57). Interacts with cog in affecting recombination in his-3 and crossing over between his-3 and ad-3 (45, 336). Used in conjunction with translocation T(TM429) his-3 to demonstrate that cog+ acts only in cis in affecting recombination between sites in his-3 (336). (See cog.) Recessive rec-2 alleles from other lineages were called rec-4, rec-5, or rec-w until identity was demonstrated (333).
rec-3 : recombination-3
IL. Between acr-3 (1% or 2%) and arg-3 (2%; 6%) (334, 337).
Presence of allele rec-3+ reduces recombination within the loci his-2 (IR) and am (VR) but not within gul-1, which shows <0.3% recombination with am (337, 1940, 1941). Crossing over is also reduced in the interval between sn and his-2 (338) (Fig. 57). Three alleles are known: rec-3, rec-3L, and rec-3+ (334). A recessive allele from another lineage was initially called rec-x (333).
rec-4, rec-5, rec-w: recombination-4, -5, -w
All are rec-2 alleles.
rec-x : recombination-x
A rec-3 allele.
rec-z : recombination-z
A rec-1 allele.
Restless (Transposable Element)
An Ac-like DNA-transposable element introduced into Neurospora from the asexual fungus Tolypocladium inflatum (1042, 1043).
rg-1 : ragged-1
IR. Between T(AR173)R (hence of his-2) and lys-4 (1%; 7%) (492, 1548, 1578, 1583).
Spreading dense colonial growth with poor conidiation (1548). Increased hyphal branching, bumpy mycelial surface. Altered phosphoglucomutase (EC 5.4.2.2) (isozyme I). Accumulates glucose-1-phosphate (243). Cell-wall composition (287). Normal levels of NADPH (233) and linolenic acid (241). Photograph (235). The double mutant rg-1 cr-1, which grows on agar as small, discrete conidiating colonies suitable for velvet replication (1256), is not homozygous-fertile unlike sn cr, which it resembles phenotypically. For examples of applications, see refs. (1830) and (1979). Allele R2357 called er: erupt. Allele S4357 called col-7.
rg-2 : ragged-2
IR. Linked to mat (15%), rg-1 (0/650) and left of lys-4 (12%) after introgression into Neurospora crassa (1546).
Altered phosphoglucomutase (EC 5.4.2.2) (isozyme II) (1357). Morphology is similar to rg-1 (1359). Found in a Neurospora sitophila cross that was heterozygous for an rg-1 allele from N. crassa. Interpreted from its behavior in N. sitophila to be unlinked to rg-1 or to su(rg-2) (1359).
rgb-1 : B-regulatory subunit-1
IL. Linked to leu-4, mat, arg-3 (0/18) (1447).
Cloned and sequenced: GenBank AF077355.
Encodes the B-regulatory subunit of the type 2A phosphoprotein phosphatase complex (PP2A) (2273). Intron I, which is 5' to the presumed translocation initiation codon, contains an upstream ORF encoding 34 amino acids. Intron VI undergoes alternative splicing. RIP-induced inactivation of rgb-1 produced progeny that grow slowly, have abnormal hyphal morphology, are female-sterile, and produce abundant arthroconidia. Microscopic and genetic analyses indicate that rgb-1 is a regulator of the budding subroutine of macroconidiation. For a review of protein phosphatases in filamentous fungi, see ref. (535).
rgr : resistant to glutamine repression
Unmapped.
Arginine catabolism is resistant to glutamine repression (749).
rhd : rehydrin
VII? Shows sequence homology to a region in VII contigs (1041).
Cloned and partially sequenced: EMBL/GenBank AI398918, AI416419; EST W09E10, W06E2.
Putative structural gene for rehydrin (thiol-specific antioxidant, peroxiredoxin, dormancy-associated protein, non-selenium glutathione peroxidase, EC 1.11.1.7). The preceding ESTs define the 5' and 3' ends of a transcript with homology to a region in linkage group VII contigs (EMBL/Genbank AC006498; 1041). The sequence shows high homology to the rehydrin gene, which is highly conserved from cyanobacteria to higher plants and animals. Consistent with the probable involvement of an rhd gene in dormancy, the sequence is found in a subtracted conidial cDNA library (1448).
rhy-1 : rhythm-1
Unmapped.
Shows circadian rhythmic conidiation with period length 23 hr at 26ºC. Temperature-compensated. Arrhythmic at 30ºC. Ascospores germination is very poor. Obtained by insertional mutagenesis (352a).
rib : riboflavin
Only two rib loci are known in Neurospora, compared to six that have been assigned to biosynthetic steps in Saccharomyces (522). Supplemented medium should be shielded from light to avoid the destruction of riboflavin.
rib-1 : riboflavin-1
VIR. Between ad-1 (3%; 6%), Cen-VI (1%), glp-4 (4%), T(AR209) and pan-2 (3%) (915, 1986, 2160).
Requires riboflavin (1361). Used to examine the role of flavin as a photoreceptor for carotenogenesis and for the phase-shifting and suppression of circadian conidiation (1513). Allele 51602 is heat-sensitive (34 vs 25ºC), but allele C106 is not (717).
rib-2 : riboflavin-2
IVR. Between T(4342)L and chol-1. Probably left of pyr-3 (1/24 asci) (718, 1578, 1580).
Requires riboflavin (718). Used to demonstrate the role of flavin as a photoreceptor for the phase-shifting of circadian conidiation and carotenogenesis (1513).
Ribosomal RNA
Genes specifying 5.8S, 17S, and 25S ribosomal RNA (but not 5S) are located in the nucleolus organizer region. See rDNA, NOR, and Fsr.
rip-1 : ribosome production-1
IIR. Between trp-3 (6%; 9%); un-15 (1%) and Tip-IIR (1582).
Cloned (1818).
Scored as an irreparable heat-sensitive mutant (34 vs 25ºC). Attains 2% of the wild-type linear growth rate at 35ºC and 80% at 25ºC (1755). Conditional defect in the accumulation of 25S rRNA and, hence, 60S cytosolic ribosomal subunits (1219, 1221, 1222, 1755). Defective ribosome biosynthesis is attributed to a defect in rRNA processing (1222). At restrictive temperature, conidia survive and apolar growth results in highly multinucleate spherical cells, with no hyphae on glycerol complete agar medium (1546, 1751). Messenger RNA synthesis is normal at 37ºC. Good fertility, growth, and viability at 25ºC make rip-1 preferable to un-15 as a marker for the right end of II. The original strain carrying both rip-1 and inl was called 4M(t), and the rip-1 allele was designated 1(t).
ro : ropy
Morphologically similar mutants, so named because the hyphae form ropelike aggregates that grow up the tube wall from agar slants, producing conidia in dense clumps at the top (1548). Hyphae are excessively branched and curled (248, 719). Microscopic analysis reveals defects in conidial germination, placement of septa, and mitochondrial morphology (1348). ropy mutants function as fertilizing parents, but are female-infertile unless complemented in a heterokaryon with ro+. None of the ropy genes has been found to be essential for viability [see, for example, ref. (2093)]. Nuclear distribution in hyphae is abnormal as a result of defects in a microtubule-associated motor (248, 1634, 1729). Mutations at various ropy loci can be selected as partial suppressors of cot-1 [248, 1637; reviewed in ref. (2258)]. Some combinations of allelic mutations show complementation between alleles, whereas some mutations at different ropy loci fail to complement one another (248). Unlinked noncomplementation most often involves ro-1 or ro-3.
ro-1 : ropy-l
IVR. Linked to pan-1 (0/394), hence, between ad-6 and cot-1 (1255).
Cloned and sequenced: Swissprot DYHC_NEUCR, EMBL/GenBank L31504, PIR B54802, GenBank NEURO1DHC; Orbach-Sachs clone X1B06.
Morphology is typical ropy. Encodes the heavy chain of cytoplasmic dynein, a minus-end-directed motor molecule. Some combinations of alleles show complementation and may fail to complement some alleles at other ropy loci (248). Used to examine the kinesin-dynein (kin; ro-1) double mutant, which is defective in oppositely directed microtubule motors, and to compare it with the single mutants (1871). Photographs (507, 1626). The amount of cell-wall peptides is reduced (2249). Growth is limited on glycerol medium (419).
ro-2 : ropy-2
IIIR. Between trp-1 (2%, l4%) and T(D305)L, phe-2 (5%) (18, 1578, 1585). Adjoins crps-7 (2155).
Cloned and sequenced: EMBL/GenBank U23425; Orbach-Sachs clone X20H05.
Encodes a novel 80-kDa protein that has two cysteine-rich motifs that resemble zinc-binding LIM or RING domains thought to mediate protein-protein interactions. Mutations obtained by RIP inactivation are defective in nuclear migration and are blocked in conidiation (2155).
ro-3 : ropy-3
IIL. Between pyr-4 (1% or 2%) and T(NM149), thr-2 (6%; 25%) (1580, 1585).
Cloned and sequenced: Swissprot DYNA_NEUCR, EMBL/GenBank L48661, GenBank NEURO3DYN; Orbach-Sachs clone X11C06.
Encodes the largest subunit of the dynactin complex (P150Glued), which is not essential for viability but is required for normal nuclear distribution (2093). Some combinations of alleles show complementation and may fail to complement some alleles at other ropy loci (248). Photograph (248). Growth is limited on glycerol medium (419). The Spitzenkörper is undetectable or difficult to detect by phase contrast (1727). Called cfl on the map in ref. (1585).
ro-4 : ropy-4
VR. Linked to pab-2 (0/407) and carried in the same plasmid (1729). Between ad-7 (4%), T(EB4)R, and inv (5%) (321, 1578, 1582, 1592).
Cloned and sequenced: Swissprot ACTZ_NEUCR, EMBL/GenBank L31505, U14008, PIR A54802, PS00132, GenBank NEURO4CEN; Orbach-Sachs clone X9B02.
Structural gene for actin-related protein I (centractin). Mutations obtained by RIP or gene disruption are viable (1729). Photograph (1634). Growth is limited on glycerol medium (419). The amount of cell-wall peptides is reduced (2249). Mutants R2428 and R2520, originally called ro-5 and ro-8 (719, 1383), are allelic with ro-4 (allele B38) (1592).
ro-5 : ropy-5
Used for a ro-4 allele (1592).
ro-6 : ropy-6
IR. Between T(4540)L nic-2 (0/95) and T(4540)R, thi-1 (719, 1578, 1582).
Hyphae form ropelike aggregates (719).
ro-7 : ropy-7
IIL. Linked to pi (0/75), left of cys-3 (11%) (719, 1582).
Cloned and sequenced: Orbach-Sachs clone G18A08.
Encodes a novel 70-kDa actin-related protein (1347). The large subunits of cytoplasmic dynein and dynactin accumulate at spindle pole bodies in the mutant (1347). Growth is inhibited on glycerol medium (419). Female sterile, contrary to a misprint in ref. (1584).
ro-8 : ropy-8
A ro-4 allele.
ro-9 : ropy-9
Misnamed? The existing stock is probably an allele of dapple (1567). See da.
IIL. Linked to thr-3 (0/63), arg-5 (8%). Right of T(NM149) (719, 1567).
The validity of the available strain, R2526, is suspect. It is not morphologically ro and is indistinguishable from da. Conidia of R2526 are normal sized, unlike tng. The mutant may have been misidentified and misnamed (unlikely), the original may have been a double mutant from which the ropy component was lost, or a stock may have been mislabeled (1567).
ro-10 : ropy-10
IL. Between Tip-IL and cyt-21 (1%), fr (18%), In(SLm-1)L (114, 976, 1578, 1582).
Cloned and sequenced: EMBL/GenBank AF015561; Orbach-Sachs clones X7A06, G17G08.
Encodes a novel 24-kDa protein that may be necessary for dynactin stability (1347). Ascospores from rearrangement Slm-1 ´ Normal include a class that is deficient for a short terminal segment containing ro-10. The deficiency progeny germinate and die unless they are rescued in a heterokaryon. The rescued deficiency can be transmitted through a cross (114).
ro-11 : ropy-11
IIIR. Linked to dow (0/70). Between T(AR17)L and T(AR17)R (1347, 1349).
Cloned and sequenced: EMBL/Genbank AF015560; Orbach-Sachs clone G25B06.
Encodes a novel, nonessential 75-kDa protein involved in nuclear distribution (1347, 1349). Obtained by RIP in a cross of Dp(AR17) ´ Normal (1349).
rol : ropy-like
This name has been given to mutants that resemble ropy in growth habit on slants, but that differ from ropy in having hyphae that are not seen microscopically to be curled (719).
rol-1 : ropy-like-1
IV. Linked to pdx-1 (0/88) (719).
Forms mycelial aggregates similar to those of ro mutants, but with hyphae not curled microscopically.
rol-2 : ropy-like-2
VII. Linked to met-7 (<1%), wc-1 (3%) (1582).
Forms mycelial aggregates similar to those of ro mutants, but with hyphae not curled microscopically.
rol-3 : ropy-like-3
VR. Between ilv-1 (2%) and cot-4 (5%) (1383).
Ropylike hyphal morphology (1383). Photograph: Fig. 16 in ref. (719).
ros : rosy
Allelic with al-3.
Rsp : Round spore
The symbol R has precedence.
rug : rug
IVR. Between pyr-2 (3%) and T(NM152)R, cys-4 (10%) (1255, 1414, 1578, 1585).
Spreading colonial morphology, with conidiating tufts. Grows better on sucrose minimal medium than on glycerol complete (1548). Formerly called mat. The name was changed to rug so that mat could be used as the symbol for mating type, in conformity with usage in other fungi (745).
Return to the 2000 Neurospora compendium main page