s : suppressor of pyr-3
Changed to arg-12s.
sar-1 : surfactant resistant-1
I. Near mat (36).
Resistant to the surface-active agents dequalinium chloride, cetyltrimethylammonium bromide, and benzalkonium chloride. Resistant growth follows an adaptive lag phase. Wild type Em A (FGSC 627) and related mat A strains carry a mat-linked sar gene that may be sar-1 (36).
sar-2 : surfactant resistant-2
Unmapped. Independent of sar-1 (36).
Resistant to surface-active agents. Phenotypically similar to sar-1 (36).
sar-3 : surfactant resistant-3
I. Near mat (36).
Phenotypically distinct from sar-1. Evidence of nonallelism is not given (36).
sat : satellite
VL. At Tip-VL, distal to the NOR (1307, 1578). Linked to lys-1 (35%) (118).
Microscopically visible terminal satellite (trabant). Best seen at pachytene in orcein- or acriflavine-stained preparations as a tiny dot on the surface of the nucleolus at one end of the second longest chromosome (1307). Photographs (118, 1598, 1599). Neurospora crassa laboratory strains differ in the presence (sat+) or absence (sat-) of a satellite. No satellite has been found in Neurospora intermedia or other Neurospora species. The satellite was used as a marker in assigning the NOR to the left arm of linkage group V (118).
sbr : small brown
Unmapped.
Cloned and sequenced (2153).
The sequence suggests a function in transcriptional regulation. Forms compact, slow-growing colonies that darken with age. No conidia or conidiophores are produced. Sausage-shaped cell compartments produce large spherical buds. Ascospores do not germinate. The mutant allele was obtained by insertional inactivation using an hph construct, and the hph tag was used to clone sbr from a genomic DNA library (2153).
sc : scumbo
IIIR. Between acr-2 (3%; 6%), Cen-III (4T/230 asci) and ser-1 (4%) (103, 1546). Linked to thi-4 (1/280), spg (0++/179) (931, 1587).
Irregular flat, spreading growth with knobby protrusions and abnormal conidiation, but no free conidia (Fig. 58). Mycelium usually appears yellowish rather than orange. Female fertile. Asci are nonlinear in crosses homozygous for sc (1623, 1624, 1969). The ascus abnormality is recessive. Cell-wall analysis (287). Cell-wall peptides are reduced in amount (2249). Resistant to vinclozolin and other dicarboximide fungicides (789). Allele R2503 was initially called col-14, and allele R2386 was called smco-2. Morphology altered by mod(sc).

FIGURE 58 The mutant sc: scumbo. Bar length = 1 mm. Scanning EM photograph from M. L. Springer.
scon-1 : sulfur control-1
VR? Probably linked to his-6, but inviable white spores are prevalent in sconc crosses and the presence of a translocation (and therefore pseudo-linkage) cannot be ruled out. Difficult to map because of infertility (1322).
The wild-type allele specifies a negative regulator in the sulfur catabolic regulatory circuit (Fig. 24). Allele scon-1c makes production of these enzymes constitutive. Prototrophic. Non-repressible for sulfate permease and therefore resistant to chromate under conditions of repression (e.g., high methionine) when it would be expected to be sensitive. The effect in a heterokaryon is restricted to its own nucleus (259). Hypostatic to regulatory mutant cys-3 (539). Ascospores and conidia germinate poorly (259, 1822). For reviews of the sulfur system, see refs. (1283) and (1287).
scon-2 : sulfur control-2
IIIR. Between trp-1 and Fsr45 (1334). Linked to tyr-1 (18%), leu-1 (26%) (538).
Cloned and sequenced: EMBL/GenBank U17251.
Encodes a β-transducin; contains an evolutionarily conserved F-box domain that is necessary for the regulation of cys-3 (1110, 1111, 1516). The wild-type allele specifies a negative regulator in the sulfur catabolic regulatory circuit (Fig. 24). Prototrophic. Resembles scon-1c in phenotype, but action is not nucleus limited. RNA transcription that requires CYS-3 occurs during sulfur-derepression, but expression of CYS-3 under sulfur repressing conditions is not sufficient for scon-2 expression, though it is sufficient for ars expression (1287, 1517). For reviews of the sulfur system, see refs. (1283) and (1287).
scot : spreading colonial temperature sensitive
VR. Between al-3 (7%) and his-6 (11%) (1583).
Not distinguishable from scot+ at 25ºC under most conditions, but above 34ºC growth on solid medium is spreading colonial, with delayed, reduced conidiation, whereas growth in liquid medium is pelleted. Readily scorable on glycerol complete medium at 39ºC. Photograph (980). Present in Beadle-Tatum and Rockefeller-Lindegren wild types and in numerous derivatives from them (1583). The same gene probably was discovered independently by Fincham (637) and by Emerson (608), studied by Pao (1531), and called t: thermophobic. Strains containing t showed start-stop growth on growth tubes, with growth distance depending on the carbon source (sucrose vs lactose or galactose) and concentration. The effect is greater on poorly utilized carbon sources and on the poorest, galactose, can be seen even at 25ºC (637). scot might be responsible for the effect of temperature on moe mutants that originated in the RL wild type (719) and (in part) for the erratic growth of heterokaryons involving Wilson-Garnjobst het-tester strains (980).
scp : serine carboxypeptidase
IR. Linked near Fsr-18 (1036).
Cloned and sequenced: Orbach-Sachs clone G3G11.
Encodes serine carboxypeptidase. Homologous to kex-1 of Saccharomyces. Inactivation of scp results in reduced fertility in mat A strains, but scp mat a strains behave normally (1036). For other genes expressed differentially in the two mating types, see ccg-4, eat-1, eat-2, mfa-1.
scr : scruffy
IIR. Linked to arg-12 (2%), probably to the left (1582).
Semicolonial growth with few aerial hyphae, dichotomization of hyphal tips, and reduced conidiation. Heat-sensitive, growing poorly with no conidia at 39ºC, more like wild type at 25ºC. Asci homozygous for scr are abnormal, with altered arrangement of ascospores. Some asci are linear. Occasional asci have fewer than eight ascospores, and some of these are large. The ascus abnormality is recessive (1623).
sdh-1 : succinate dehydrogenase-1
I. Linked to mat (0/13) (590).
Succinate dehydrogenase (SDH) activity is 18% that of wild type. Succinate oxidase activity is low. Selected by failure to reduce nitrotetrazolium blue in the presence of succinate and phenazine methosulfate in an overlay after inositol death enrichment on acetate (590). Deficient in the HiPIP iron-sulfur center of the SDH complex (589).
sdv : sexual development
Subtractive hybridization was used to identify genes expressed specifically under conditions that induce sexual development. With two exceptions, sdv-10 and sdv-15, the sdv genes listed here showed no detectable phenotype after being disrupted by RIP. (sdv-10 and -15 were renamed asd-1 and asd-3.) sdv-1, -2, and -3 are clustered on cosmid 4.3A, sdv-4 and -5 are on cosmid 5.6F, sdv-6 is on cosmid 8.9A, sdv-7, -8, and -9 are on cosmid 12.2E, sdv-10 and -15 are adjacent on cosmid 12.7B, sdv-11 and -12 are on cosmid 20.11A, and sdv-13 and -14 are tightly linked on cosmid 28.12G. Mapping was by RFLP (1450).
sdv-1 : sexual development-1
IVL. Between Tel-IVL (12T, 0NPD/18 asci) and Cen-IV, aod-1 (0, 1, or 2T/18 asci) (1447).
Cloned: on cosmid 4.3A with sdv-2 and sdv-3.
Expressed specifically under conditions favoring sexual development (1450).
sdv-2 : sexual development-2
IVL. Between Tel-IVL (12T, 0NPD/18 asci) and Cen-IV, aod-1 (0, 1, or 2T/18 asci) (1447).
Cloned: on cosmid 4.3A with sdv-1 and sdv-3.
Expressed specifically under conditions favoring sexual development (1450).
sdv-3 : sexual development-3
IVL. Between Tel-IVL (12T, 0NPD/18 asci) and Cen-IV, aod-1 (0, 1, or 2T/18 asci) (1447).
Cloned: on cosmid 4.3A with sdv-1 and sdv-2.
Expressed specifically under conditions favoring sexual development (1450).
sdv-4 : sexual development-4
IR. Between al-2 (1T/18 asci) and arg-13 (5T, 2 NPD/18 asci). Linked to vma-11 (0 or 1T/18 asci) (1447).
Cloned: on cosmid 5.8F with sdv-5.
Expressed specifically under conditions favoring sexual development (1450).
sdv-5 : sexual development-5
IR. Between al-2 (1T/18 asci) and arg-13 (5T, 2NPD/18 asci). Linked to vma-11 (0 or 1T/18 asci) (1447).
Cloned: on cosmid 5.8F with sdv-4.
Expressed specifically under conditions favoring sexual development (1450).
sdv-6 : sexual development-6
VR. Linked to crp-4 (0T/18 asci). Right of inl (9T/18 asci), tom70 (4T/18 asci), Fsr-20 (1T/18 asci) (1447).
Cloned: on cosmid 8.9A.
Expressed specifically under conditions favoring sexual development (1450).
sdv-7 : sexual development -7
IVR. Right of trp-4 (5T/18 asci), crp-3 (1T/18 asci) (1447).
Cloned: on cosmid 12.2E with sdv-8 and sdv-9.
Expressed specifically under conditions favoring sexual development (1450).
sdv-8 : sexual development-8
IVR. Right of trp-4 (5T/18 asci), crp-3 (1T/18 asci) (1447).
Cloned: on cosmid 12.2E with sdv-7 and sdv-9.
Expressed specifically under conditions favoring sexual development (1450).
sdv-9 : sexual development-9
IVR. Right of trp-4 (5T/18 asci), crp-3 (1T/18 asci) (1447).
Cloned: on cosmid 12.2E with sdv-7and sdv-8.
Encodes glycogen phosphorylase (EC 2.4.1.1) (1446). Expressed specifically under conditions favoring sexual development (1450).
sdv-10 : sexual development-10
Changed to asd-1.
sdv-11 : sexual development-11
IVR. Between pyr-1 (4T/18 asci) and trp-4 (3T/18 asci) (1447).
Cloned: on cosmid 20.11A with sdv-12.
Expressed specifically under conditions favoring sexual development (1450).
sdv-12 : sexual development-12
IVR. Between pyr-1 (4T/18 asci), and trp-4 (3T/18 asci) (1447).
Cloned: on cosmid 20.11A with sdv-11.
Expressed specifically under conditions favoring sexual development (1450).
sdv-13 : sexual development-13
VII. Linked to ars-1, Cen-VII (0T/18 asci) (1447). On the same plasmid with sdv-14 (1450).
Expressed specifically under conditions favoring sexual development (1450).
sdv-14 : sexual development-14
VII. Linked to ars-1, Cen-VII (0T/18 asci) (1447). On the same plasmid with sdv-13 (1450).
Expressed specifically under conditions favoring sexual development (1450).
sdv-15 : sexual development-15
Changed to asd-3.
sen : senescent
VR. Between his-1 (4%) and al-3 (13%) (1441b).
Resembles the mutant natural death in progressive deterioration of mycelial growth followed by death. Recessive. The mutant gene was extracted from a heterokaryotic wild-collected strain of Neurospora intermedia by plating microconidia that were obtained as described in ref. (1528). Introgressed into Neurospora crassa by nine backcrosses. Stocks are maintained in a heterokaryon with the am1 ad-3B cyh-1 helper strain, from which sen is extracted by crossing to wild type (1441b).
ser-1 : serine-1
IIIR. Between sc (4%) and pro-1 (1%; 114 kb; 9%), ace-2 (5%) (448, 940, 1546).
Cloned: pSV50 clones 14:1C, 22:4D, 25:3G (448).
Slightly deficient in serine hydroxymethyltransferase; raised levels of 10-formyltetrahydrofolate synthetase. Lacks the ability to incorporate C1 units from glycine. Extracts lack detectable methylfolates. Negligibly deficient in 5,10-CH2-H4PteGlu reductase (411). Uses serine or glycine (940). Inhibited on organic complete medium. Ascospores from ser-1 ´ ser-1 do not blacken or do so only after a long delay (1592). Morphologically normal; the abnormal morphology reported at elevated temperatures in ref. (1592) was due to scot (1583).
ser-2 : serine-2
VR. Between met-3 (4%) (824) and T(EB4)L, cot-2 (5%; 8%) (321, 322).
Uses serine (556), but not glycine (1546). Does not grow on hydrolyzed casein (556). Inhibited by thienylserine (99).
ser-3 : serine-3
IL. Between cys-5 (1%) and In(NM176)L, un-3 (<1%) (1578, 1592, 2129).
Uses serine and (less well) formate. Also grows fairly well on a combination of adenine, methionine, tryptophan, and lysine. Does not grow on casein hydrolysate (556). No or little response to glycine (1546). Deficient in phosphoserine phosphatase activity (EC 3.1.3.3) (139). Inhibited by leucine. Used to examine one-carbon metabolism (989). Scorability good, but vigor and leakiness vary markedly in different isolates. Homozygous crosses give mostly white ascospores (1546, 1592).
ser-4 : serine-4
IVR. Right of arg-2 (<1%) (1300, 1301).
Uses serine or glycine. Incompletely blocked. Not deficient for any of the enzymes involved in serine synthesis from 3-phosphoglyceric acid or glyceric acid. The intracellular pool is deficient in serine, glycine and alanine, and it accumulates threonine and homoserine (1301). Produces abundant L-amino acid oxidase, but no tyrosinase, while growing slowly on minimal medium (1301, 2139). Allele DW110 called P110.
ser-5 : serine-5
IIIR. Linked to trp-1 (1%), ser-1 (12%) (1302).
Uses serine or glycine. Incompletely blocked (1302).
ser-6 : serine-6
VIL. Between nit-6 (10%) and het-8 (8%), ad-8 (16%) (1034, 1172, 1582).
Responds to serine. Slight response to glycine. Extremely leaky. Scorable by slow, sparse conidiation on minimal slants or auxoanographically (1546). Can be crossed without supplement on minimal crossing medium. Allele DK42 was obtained as a putative leucine regulatory mutant (1034), but a regulatory role is dubious (793). Called DK42 (1034). Probably allelic with T(OY325) ser, which has a breakpoint left of lys-5 (11%) in VIL and shows 0/28 recombination with DK42, which it resembles in leakiness. OY325 grows more profusely than DK42 on minimal slants (1582).
sf : slow-fine
I. Linked to mat (3%), cy (3% in regular perithecia) (1366, 1582).
Originally detected microscopically. Growth from ascospores or from conidia is slow at first. Fully grown cultures appear morphologically normal. Asci are irregular in some perithecia (1366). Scorable by slow growth on slants 3 or 4 days after ascospore germination. Hyphae initially grow clockwise on an agar surface (1582).
sfo : sulfonamide dependent
VII. Linked to Cen-VII (<1%) (604). Between thi-3 (6%) (1713) and hlp-1 (1%; 9%) (879).
Requires sulfonamide at 35ºC and is stimulated by sulfonamide at lower temperatures (605). Overproduces p-aminobenzoic acid; hence, growth is inhibited by exogenous PABA (2293). In a sfo; pab-1 double mutant on sulfonamide-supplemented minimal medium, PABA is stimulatory at very low concentrations but inhibitory at higher (605). In a sfo; met-1 double mutant on sulfonamide- supplemented minimal medium, methionine is stimulatory at very low concentrations but inhibitory at higher (2294). Best tested on solid minimal growth tubes at 35ºC. Suppressor mutations are frequent (604).
sg : spontaneous germination
Unmapped.
Ascospores germinate spontaneously, without heat shock. A component of the multiple-mutant combination responsible for the cell-wall-deficient “slime” phenotype. Usually associated with very poor vegetative growth. Possibly more complex than a single gene (607). (Ascospores carrying the fluffy mutation often germinate spontaneously.)
sh : shallow
VR. Between ilv (4%; 7%) and md (3%) (569).
Hyphae spread in a fanlike array and do not penetrate deeply into agar (1585). Photographs (569, 719).
shg : shaggy
IIIR. Linked to trp-1 (7%), acr-6 (0/368). Recombines with vel, col-13 (932, 1582).
Growth is slow. Conidia are formed on the irregular aerial hyphae that are most abundant high in an agar slant. Mutants acr-4 and acr-6 originated in shg. The acr-4 gene confers resistant to acriflavine only in combination with shg (932). Called mo(KH160).
sit : siderophore transport
The uptake of exogenous ferricrocin and coprogen is defective. The five known sit mutations appear from intercrosses to be nonallelic (362). Double mutants prove that sit-2, sit-3, and sit-5 are at different loci. Differences are present in morphology, pigment, speed of conidial germination, female fertility, and binding of siderophores. Map locations are unknown. The mutants were selected using aga; arg-5; ota, which is blocked in the known pathways to ornithine (463) and thus depleted of siderophores (2229) (Fig. 10). (Because exogenous arginine strongly represses ornithine synthesis, use of the aga single mutant in the presence of arginine was proposed for future experiments.) The five mutants, which grow well on minimal medium, take up [14C]phenylalanine but are unable to take up [3H]ferricrocin. Methods and characteristics of the mutants were summarized when work was suspended in 1983 (362). The conditional germination-defective mutant moe-1 (JS134-9) is partially curable by siderophores and may have an altered plasma membrane attachment site (360). For background on the Neurospora siderophore uptake system, see ref. (2228). For the molecular biology of fungal siderophores, see ref. (1162).
sit-1 : siderophore transport-1
Unmapped.
Transport of siderophores is severely reduced. Binding is defective. Germination is slow. A double mutant with sit-2 is available, and this also shows slow germination (359, 362).
sit-2 : siderophore transport-2
Unmapped.
Transport of siderophores is severely reduced. Binding is defective. Conidia tend to stay connected in twos and threes. The color is deep orange. Double mutants are available with sit-1 (FGSC 4226, 4227), sit-3 (FGSC 4219, 4920, 4921, 4922), and sit-5 (FGSC 4223, 4224, 4225). These all have the morphological characteristics of the sit-2 parent (359, 362).
sit-3 : siderophore transport-3
Unmapped.
Transport of siderophores is severely reduced. Binding is normal. Double mutants are available with sit-2 and sit-5 (FGSC 5230, 5231) (359, 362).
sit-4 : siderophore transport-4
Unmapped.
Transport of siderophores is severely reduced. Binding is normal. A double mutant with sit-5 is available (FGSC 4228, 4229) (359, 362).
sit-5 : siderophore transport-5
Unmapped.
Transport of siderophores is severely reduced. Binding is normal. Double mutants are available with sit-2, sit-3, and sit-4 (359, 362).
sk : skin
VIIR. Between nt (7%; 17%) and Tip-VIIR (1548).
Leathery, rapid surface growth, nonconidiating for most alleles (1548). “Mucilaginous substrate hyphae” (2117). Ascospores of sk constitution are slow to mature, but good allele ratios are obtained from crosses held for 3 weeks at 25ºC. Female sterile. Allele P1718 is exceptional in making aerial hyphae and conidia, best on complete medium. Allele R2466 called mo-3; alleles Y6821, R2408, and R2529 called moe-1 (719, 1582). Not to be confused with Sk: Spore killer.
Sk : Spore killer
When a chromosomal Spore killer element is heterozygous in a cross, it results in the death of ascospores that do not carry the element. In crosses heterozygous for a killer element, each ascus contains four viable black ascospores and four inviable unpigmented ascospores (Fig. 59). The surviving spores in 4:4 asci are all Sk. Ascospores are not killed in crosses homozygous for the same Sk element (2125). Three different killer elements are known, all from natural populations. The best studied are Sk-2 and Sk-3, which were found in Neurospora intermedia and crossed into Neurospora crassa for mapping and analysis. Both are gene complexes (“haplotypes”) that, when heterozygous, block meiotic recombination over an interval ~30 map units long on linkage group III. Sk-2 and Sk-3 are distinguished by the specificity of killing and by the specificity of their response to genes that confer resistance to killing. In crosses where killing occurs, meiosis and two postmeiotic mitoses appear to be normal, as judged by light microscopy. Sensitive ascospores first appear abnormal after one nuclear division in the ascospore. No killing occurs in crosses homozygous for a killer nucleus of the same type, but Sk-2 ascospores are subject to killing by Sk-3 and vice versa. Likewise, genes conferring resistance to killing by one type do not confer resistance to the other. Less than 1% of ascospores are viable in crosses of Sk-2 ´ Sk-3 (2121, 2125). A sensitive nucleus that otherwise would be killed survives if it is included in the same ascospore with a killer nucleus (1674). If Sk-2 and Sk-3 nuclei are included in the same ascospore, the ascospore is not killed and both nuclei survive (1673). Because the four ascospores of Neurospora tetrasperma normally are heterokaryotic, sensitive nuclei are sheltered from killing in ascospores from crosses heterozygous for Spore killer. Crosses with Sk-2 and Sk-3 in N. tetrasperma can therefore be used to examine differences in structure and gene order within the recombination block (1686). Because the known Sk elements segregate at the first division of meiosis, heterozygous crosses can be used to map centromeres using physically unordered asci ejected from the perithecia (1602). For reviews, see refs. (1681), (1682), and (2126). For genes conferring resistance to killing, see r(Sk-2), r(Sk-3), and ref. (2124). For reference strains and tester strains for identifying the various killers and genes conferring resistance to killing, see refs. (2127) and (2128). Spore killer (Sk) is not to be confused with skin (sk), a recessive gene in VIIR.
Sk-1 : Spore killer-1
Unmapped. Less that 1% second-division segregation (2125).
Resembles Sk-2 in behavior (1674, 2125). Known only in Neurospora sitophila. Attempts to transfer Sk-1 into Neurospora crassa or Neurospora intermedia have failed. The frequency of Sk-1 in collected strains of N. sitophila is high relative to that of Sk-2 and Sk-3 in natural populations of N. intermedia (1674, 2125)
Sk-2 : Spore killer-2
IIIL,R. A haplotype. Between cum (2%) and his-7 (21%) (280). When Sk-2 is heterozygous, crossing over is suppressed in what is normally a 30-unit interval extending from r(Sk-2) through leu-1 and spanning Cen-III (2125). Markers acr-7, acr-2, and leu-1 were inserted into the Sk-2 recombination block selectively at frequencies of 10–5 or 10–6. When these were used in crosses homozygous for Sk-2, crossing over in the long blocked segment was normal (280).
The four non-Sk ascospores in each ascus abort when Sk-2 is heterozygous (Fig. 59). Asci with second-division segregation patterns are rare or absent. Strains carrying Sk-2 have been found at only four sites: Borneo (2), Java, and Papua-New Guinea. Most wild Neurospora intermedia strains are sensitive to killing by Sk-2, but resistant strains are common in some geographic areas (2121). See r(Sk-2). Two Sk-2-resistant strains have been found in Neurospora crassa, even though killer strains have never been identified in that species.
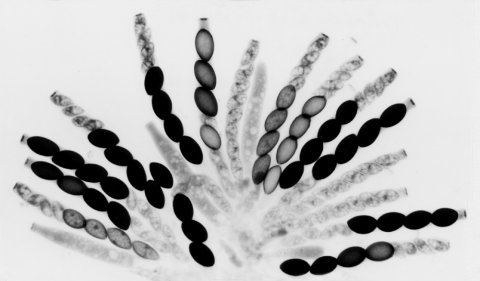
FIGURE 59 Maturing asci from a cross heterozygous for Spore killer-2. Most asci contain four normal large spores and four small aborted spores. The few asci that do not show the 4:4 pattern are still immature. All the maturing ascospores are Sk-2. Sk-2 segregates at the first meiotic division in all asci, indicating that there is no crossing over between the Spore killer complex and the centromere. Photograph by N. B. Raju. From ref. (1674 ), with permission from the Genetics Society of America.
Sk-3 : Spore killer-3
IIIL,R. A haplotype. Between cum (2%) and his-7 (12%) (280). When the Sk-3 complex is heterozygous, crossing over is suppressed in the same interval as with Sk-2. Sk-3 did not then recombine with leu-1 (none in 5 ´ 106). Recombination was about 10–5 for acr-7 and acr-2 (280).
Sk-3 resembles Sk-2 in its behavior. Sk-3 ascospores are sensitive to killing by Sk-2, however, and vice versa. Sk-3 has been found only once, in Neurospora intermedia from Papua-New Guinea. Strains resistant to killing by Sk-3 are present in N. intermedia populations in the Eastern hemisphere (2121).
Sk(ad-3A)
Probably an ad-3A allele.
A multiple-mutant phenotype, lacking a cell wall and growing as protoplasts or plasmodium subject to osmotic lysis. The original strain contained at least two mutations (fz, fuzzy; sg, slow germination) in addition to the already- present markers arg-1, cr-1, al-1, and os-1. (All markers are recessive.) Three of the markers, fz, sg, and os-1, are required for a slime-like phenotype (607). A variant with altered morphology, FGSC 4761, grows more rapidly (1801). The slime strain is altered in the production and secretion of several enzymes related to sugar catabolism (1622). Cell-wall-like material may be produced in small quantity in stocks newly resolved from a phenotypically wild-type heterokaryon. slime protoplasts can be induced to fuse (1453). The ability to form heterokaryons or to function as fertilizing parent is lost after continuous growth (1883). Stocks can be maintained in phenotypically normal heterokaryons of the same mating type (607) or of mixed mating type and recovered by filtration (1456). Alternatively, stocks may be kept at -70ºC in situ on agar medium (1877), in growth medium (1867), or in dimethyl sulfoxide (429). Recombinant slime-like progeny with inserted markers can be recovered from crosses using filtration enrichment (1456). The slime strain has been used for isolating and storing plasma membrane in large quantity (1287), analyzing fatty acids in the plasma membrane (681), purifying and characterizing plasma membrane H+-ATPase (1802), extracting enzymes gently (703), isolating chitosomes (124), obtaining intact chromosomal DNAs for separation by pulsed-field gel electrophoresis (816, 1129), studying vacuoles (1275, 1800), showing that poly(A) polymerase and nuclease activities are largely located in the nucleus (1868), and creating an efficient cell-free system for protein synthesis (7, 2034). Nuclei of opposite mating type appear to be compatible in a (tol + slime tol+) heterokaryon (1456). For general methodology, see refs. (1456) and (2243). Cell-wall-free Neurospora resembling slime can be obtained following growth of an os-1 strain in the presence of polyoxin D (1879, 1881).
slo-1 : slow-l
IR. Between mat (14%) and thi-1 (2%; 5%) (1548).
Growth from conidia or ascospores is slow (1548). Conidiation lags significantly behind wild type. Morphology is normal. Not tested for cytochrome deficiency or for the ability to reduce tetrazolium.
slo-2 : slow-2
VII. Left of met-7 (2%) (1592).
Growth from conidia or ascospores is slow. Conidiation lags days behind wild type (1592, 2013). Not tested for cytochrome deficiency or for the ability to reduce tetrazolium.
smco : semicolonial
This name was introduced by Garnjobst and Tatum (719) for mutants that begin growth on agar as a small colony and sooner or later produce a flare of wild-type-appearing hyphae, with or without conidia.
smco-1 : semicolonial-1
I. Linked to mat (1%), rg-1 (0/72) (719).
Growth initially is colonial (719).
smco-2 : semicolonial-2
Allelic with sc (S7).
smco-3 : semicolonial-3
I. Linked to mat (10%), al-2 (29%). Recombines with col-7, smco-1 (719).
Growth initially is colonial (719).
smco-4 : semicolonial-4
IVR. Linked to pan-1 (8%) (719)
Growth initially is colonial (719).
smco-5 : semicolonial-5
I. Linked to mat (2%).
Semicolonial flat growth persists until 4-7 days after ascospore germination, when wild-type mycelium develops. Shows phenotypically wild-type growth when transferred, but when it is crossed by smco-5+, smco-5 ascospores are produced that repeat the cycle. Recombines with rg-1, smco-l. Not fertile with smco-3; may be allelic (719).
smco-6 : semicolonial-6
VR. Linked to asn (6%), near pyr-6. Right of met-3 (14%) (321, 1383).
Growth initially is colonial (719).
smco-7 : semicolonial-7
VR. Right of ilv-1 (2%). Linked to rol-3 (0/154), sp (12%, probably to the left), chs-1 (0/18) (1021, 1383).
Cloned and sequenced: Swissprot RAS2_NEUCR, EMBL/GenBank D16137, GenBank NEUNCR2P; Orbach-Sachs clones X9E09, X11F07, X16G03, X18B08.
Growth initially is colonial. Conidiation occurs in a crescent at the top of an agar slant. Cell-wall synthesis is apparently defective. Morphology is distinct from rol-3, which complements smco-7 (719).
The gene called ras-2 is allelic with smco-7 allele R2497, which it resembles, with which it gives 0/84 wild-type recombinants, and which it can transform to wild type (1021, 1267). Sequence shows the gene to be a member of the ras superfamily. The single-base deletion found in allele R2497 is expected to truncate the protein product (1021). The null mutant obtained by RIP grows linearly on agar medium at one-tenth the wild-type rate, has fewer aerial hyphae and conidia, and periodically forms rings of short aerial hyphae with sparse conidia.
smco-8 : semicolonial-8
IVR. Linked to pan-1 (1%; 7%), smco-4 (30%) (719).
Growth sometimes flares out at the top of an agar slant (719). Unable to grow on galactose or grows as restricted colonies (1689). Resistant to dicarboximide fungicides, especially vinclozolin (789). The amount of cell-wall peptides is reduced (2249). Complements col-1, col-8 (719).
smco-9 : semicolonial-9
IVR. Linked to pan-1 (5%), smco-4 (2%), smco-8 (13%) (719).
Morphology is partially normalized by isomaltose or starch. Altered inhibitor of branching enzyme a-1,4-glucan-6-glycosyltransferase (2). Reduced amount of cell-wall peptides (2249). Crosses homozygous for smco-9 produce nonlinear asci (similar to pk) (1969). The strain originally obtained was complex, producing two types of colonials among progeny. The component that behaved as a recessive ascus mutant was designated smco-9 (1965). Resistant to dicarboximide fungicides, especially vinclozolin (789).
sn : snowflake
IR. Between Cen-I (<1%) and os-4 (<3%), T(AR173)L (1605, 2072).
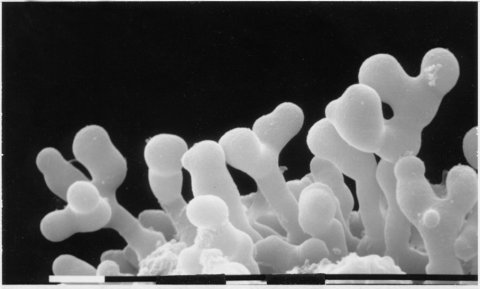
Spreading colonial growth. Linear growth is less than one-tenth wild type (34). Mutant progeny can be recognized microscopically immediately after ascospore germination by hyphal patterns that suggested the name snowflake (1365). Alleles differ in osmotic tolerance, dominance, and differentiation of conidia (2072) (Fig. 60). Large bundles of 8- to 10-nm-diameter microfilaments consisting of pyruvate decarboxylase are accumulated in vegetative cells (34, 40, 1739) (see cfp), but not during ascus development when PDC is associated with the cortical cytoskeleton (2085). The sn mutant is said not to exhibit cytoplasmic streaming (33). Meiosis and ascospore formation are normal in crosses homozygous for sn (1673). Female fertility is good. In visible appearance, sn resembles sp and cum and also cot-4 at 25ºC (1582). Used to study the development of crystalline inclusions (32). Used as a marker in a study of rec genes (338).
The double mutant sn cr-1 resembles rg cr-1 in forming small, discrete, conidiating colonies suitable for replication. sn cr-1 has the advantage over rg cr-1 of being fertile in homozygous crosses (1553). sn cr-1 has been used to select drug-resistant mutants (346). Heterokaryons between marked sn cr-1 strains are an integral part of a system developed for determining the rate of recessive lethal mutation throughout the genome (1980) (Fig. 22).
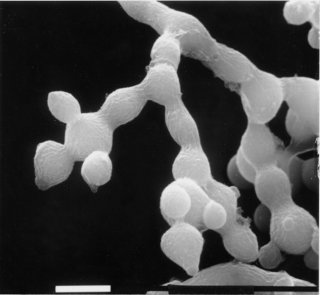
FIGURE 60 The mutant sn: snowflake (allele C136). Bar length = 10 mm. Scanning EM photograph from M. L. Springer.
snz : snooze
Allelic with pdx-1.
so : soft
IR. Between arg-13 (2%; 12%) and aro-8 (7%; 11%) (822, 1592).
Lightly pigmented. Distinctive morphology, with a lawn of fuzzy short aerial hyphae and conidia that are formed more uniformly than in wild type and closer to the agar surface, similar to peach (1548). Recurrent mutations are frequent in strains of Oak Ridge background (1011, 1014). Best scored early on short, obtuse slants. The morphological abnormality is more pronounced on sorbose-sucrose medium. Female sterile. Classified as an age mutant: the conidial life span is short under defined conditions, and the map location is near a site called age 1.3 (1394).
Hyphal fusions are not detectable microscopically. Formation of forced heterokaryons is delayed relative to wild type when ham is present in one or both strains, with a latent period of >24 hr for ham + ham+ and >80 hr for ham + ham. Once heterokaryons are formed, growth rate is reduced from 4 mm/hr for wild type to 3-3.5 mm for (ham + ham+) and 2-2.5 mm for (ham + ham) (2224). The described het genes do not affect the fusion of hyphae.
A mutant called ham: hyphal anastomosis (2224) is allelic with so (809a). The phenotype of ham was described as follows. Hyphal fusions are not detectable microscopically. Formation of forced heterokaryons is delayed relative to wild type when ham is present in one or both strains, with a latent period of >24 hr for ham ´ ham+ and >80 hr for ham + ham. Once heterokaryons are formed, the growth rate is reduced from 4 mm/hr for wild type to 3-3.5 mm for (ham + ham+) and 2-2.5 mm for (ham + ham) (2189).
sod-1 : superoxide dismutase-1
IL. Between fr (5T/16 asci) and Fsr-12, vma-4 (2 or 4T/18 asci) (1447).
Cloned and sequenced: Swissprot SODC_NEUCR, EMBL/GenBank M58687, PIR A23909, A36591, EMBL NCSOD1, GenBank NEUSOD1.
Structural gene for copper/zinc superoxide dismutase (EC 1.15.1.1) (365, 1171). A null mutant shows increased sensitivity to paraquat and oxygen as well as an increased rate of spontaneous mutation (365).
sod-2 : superoxide dismutase-2
VR. Linked to cyh-2 (0T/18 asci) (1447).
Cloned and sequenced: EMBL/GenBank AF118809.
Structural gene for mitochondrial manganese superoxide dismutase. Possibly also active in the cytoplasm (571). [The un-1 mutant has been reported deficient in SOD-2 (1392).]
sor : sorbose resistant
Sorbose is used to induce colonial growth in platings of Neurospora (231, 462). [Resolution is improved if sorbose is used in conjunction with cot-1 (332).] Numerous sor mutants have been obtained that show spreading growth on concentrations of sorbose that restrict the wild type (1075, 1076, 1757). With the exception of sor-4, scoring has usually been on minimal medium plus 0.025% filter-sterilized sorbose. Certain mutants selected for other traits may also be resistant to sorbose. For example, sor-4T9, which was selected for its ability to hydrolyze starch and which is defective in extracellular amylase, is simultaneously sorbose-resistant and osmotic-sensitive (1400). Sorbose-resistant mutants at four loci act as dominance modifiers of the ascus effect of dominant pk alleles (1757). Most sor mutants have probably not been examined for possible pleiotropic amylase or osmotic phenotypes. General references (35, 1656).
sor-1 : sorbose resistant-1
VIL. Left of ylo-1 (3%) (1076).
Defective in sorbose uptake (1075) and thereby resistant to growth restriction by sorbose (1076). Recessive (1074, 1077). Symbol changed from sor-A (1668).
sor-2 : sorbose resistant-2
VII. Linked to nt (31%) (1076).
Defective in sorbose uptake (1075) and thereby resistant to growth restriction by sorbose (1076). Recessive (1074, 1077). Symbol changed from sor-B (1668).
sor-3 : sorbose resistant-3
IIIR. Linked to ad-4 (7%) (1076).
Resistant to growth restriction by sorbose (1076). Recessive with respect to colony size. Conidiation is precocious (35). Partially recessive for percent conidial germination on sorbose test medium (1074, 1077). In heterokaryons with sor-1 or sor-2, the phenotype is intermediate between the resistant single- mutant phenotype and the sensitive wild type (1074). Symbol changed from sor-C (1668). Called sorr-17 (1076).
sor-4 : sorbose resistant-4
IL. Between mat (6%), suc (1%), In(H4250)L and arg-1 (1%, 4%), Cen-I (5%) (1400, 1592, 1988).
Cloned and sequenced (presumed allele rco-3 has been cloned and sequenced): EMBL/GenBank U54768; Orbach-Sachs clone X10E5
Resistant to growth restriction by sorbose. Osmotic sensitivity is comparable to that of os-1. Dissimilar in morphology to os-4 (1582). Growth is slow. (Putative alleles called dgr-3 and rco-3 are exceptions with growth that is not slow. rco-3 may show a lag of several hours, after which it grows at the wild-type rate on most media.) Glucoamylase and extracellular amylase are low, as is extracellular acid phosphatase activity (1400). However, dgr-3 is reported to have high non-repressible glucoamylase (35). The glucoamylase, osmotic, and sorbose-resistance properties cosegregated in 101 isolates (1400). Modifies the dominance of pk alleles (1757). The growth of gpi on glucose or sucrose is enhanced in the double mutant gpi-1; sor-4. Used to obtain mutants defective in glucose phosphate isomerase (1401). Scoring of sor-4 is clear on minimal slants with 2% sucrose + 3% sorbose after 1 and 2 days at 34ºC (1592). Alleles and probable alleles have been recognized under many different guises. Called sor(DS) (1592, 1668), Pk-mod-D (1757), T9, sor-T9 (35, 1400, 1401), gla: glucoamylase (106), amy (615). sorr-15 is a possible allele based on map position (1076). It is not clear whether sor-4 is a locus separate from pat. Allele T9 is resistant to the colonializing action of sorbose at 25ºC, but not at 35ºC (1402). Linked to the regulatory gene exo-1, which has not been tested for allelism but is stated to be on the opposite side of mat. Detected in the pat; pro-1 strain that was first used to demonstrate a circadian rhythm in fungi (1988). Mutants have been obtained by plating mutagenized conidia in medium containing starch and observing a cleared zone around colonies. Mutations called dgr, which were selected for resistance to inhibition by deoxyglucose, are also resistant to sorbose (35). Mutations designated dgr-3 appear to be allelic with sor-4 (0 + + recombinants in 400 intercross progeny) (35). A sor-4 allele formerly designated dgr-3 exhibits resistance to glucose inhibition of sexual initials (protoperithecia and melanin). A mutation called rco-3 is allelic with dgr-3 and, hence, presumably with sor-4 based on its failure to complement in (rco-3 + sor-4) heterokaryons (573). Summarized next is information obtained using the putative allele called rco-3.
The mutant previously designated rco-3 is a putative allele of sor-4 and is linked to gla-2 (<10 kb) and mat (576). It was selected as enabling higher basal expression of a con-10::hph fusion gene during vegetative growth (1241). It conidiates in liquid culture without nutrient limitation and is partially resistant to L-sorbose. The expression of conidiation-specific genes is elevated in liquid culture (1241). Sequence of the cloned gene reveals similarities to sugar transporter genes, and RCO-3 is required for normal levels of glucose-transport activity (575, 1242). Expression of qa-2 is not glucose-repressible in the mutant.
sor-5 : sorbose resistant-5
V. Linked to his-1 (1076).
Resistant to growth restriction by sorbose (1076). Mutants called sorr-14 and sorr-19 are allelic.
sor-6 : sorbose resistant-6
Probably III or VI by linkage to ylo-1 in alcoy (1582).
Resistant to growth restriction by sorbose (1076). Recessive (1075). Called sorr-6.
sor(T9) : sorbose resistant T9
A sor-4 allele.
sp : spray
VR. Between leu-5 (3%; 9%) and ure-3 (8%), am (l%; 8%; 450 kb) (219, 220, 256, 1651, 2014).
Cloned: Orbach-Sachs clone X11G10 (317).
Initially grows as a colony flat on the agar surface. Aerial mycelium then fans upward (Fig. 61) (1548). For ultrastructure and intraconidial conidia, see references in ref. (2117). Cell-wall analysis (507). Reduced amount of cell-wall peptides (2249). Morphologically similar to cot-4 (at 25ºC), sn, and cum (1582). Excellent female fertility. Used as a lawn for mating-type tests on plates (1929). Mutant morphology is corrected by calcium (533) (Fig. 61).
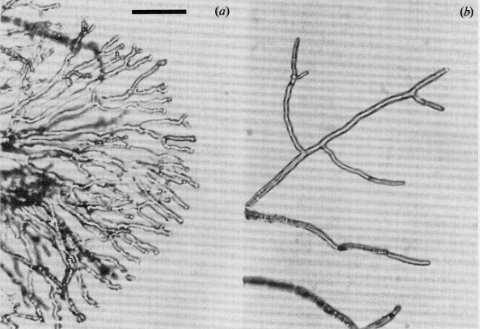
FIGURE 61Typical morphology of the mutant sp: spray on minimal medium (a) and on minimal medium plus 500 mM Ca2+ (b). The calcium supplement effectively restores wild-type morphology. Bar length-50mm. From ref. (533 ), with permission from the Society for General Microbiology.
spco : spreading colonial
This symbol and name were used in ref. (719) for mutants that begin growth on agar as a colony and do not remain restricted, but spread to cover the agar surface. Mutants of the series spco-3 to spco-15, described in ref. (719), were sometimes named as new spco genes without having been tested for allelism with already named morphological mutants having similar map locations. Growth rates and other characteristics of 11 spco mutants were described in refs. (2107) and (2108). Used to study septa and septal plugging (2109, 2110).
spco-1 : spreading colonial-1
Allelic with col-4.
spco-2 : spreading colonial-2
Allelic with wa.
spco-3 : spreading colonial-3
Allelic with spco-7.
spco-4 : spreading colonial-4
VIIL. Linked to do (<1%), nic-3 (1%, probably to the left) (1592).
Growth on agar is initially aconidial, with fine hyphae (719). Capable of conidiating on the surface of complete medium (1546). Hyphae extend faster within agar medium than on the surface, resulting in a dense hemispherical colony, most of which is embedded (2107). Septal plugging is rare in peripheral hyphae (2109).
spco-5 : spreading colonial-5
VII. Linked to col-17 (6%), nt (20%) (719).
Asci are nonlinear in crosses homozygous for spco-5 (1965). Cell-wall peptides are reduced in amount (2249). Complements col-2 and spco-4 (719).
spco-6 : spreading colonial-6
VII. Right of thi-3 (4%) (1546). Linked to spco-5 (8%), nt (20%) (719).
Complements col-17, col-2, spco-4, and spco-5 (719).
spco-7 : spreading colonial-7
VI. Linked to ad-1 (0/65). Between ylo-1 (4%) and T(AR209), trp-2 (16%; 21%) (1578, 1582).
Complements moe-2. Allele R2365 is preferred because of its excellent growth, conidiation, and fertility (1582). Allele R2365, which was originally called spco-3 and incorrectly assigned to V (719), makes small perithecia devoid of ascospores when crossed with spco-7 R2457 (1582).
spco-8 : spreading colonial-8
IV. Linked to pan-1 (23%) (719).
Spreading colonial morphology.
spco-9 : spreading colonial-9
VR. Linked to asn (6%). Right of met-3 (18%) (1383).
Multiple side branches are formed on hyphae below septa that have become occluded as a result of flooding (2110). Complements ro-5, cot-2, and smco-6. Morphologically distinct from col-9, but allelism tests were inconclusive (719, 1383).
spco-10 : spreading colonial-10
VR. Between ilv-1 (24%) and inl (5%) (21 asci) (719, 1383).
Spreading colonial morphology.
spco-11 : spreading colonial-11
IL. Between leu-3 (7%) and In(NM176)L, mat (17%). Recombines with mo-5 (18%) (719, 1578).
Conidia are occasionally produced at the top of a slant (719).
spco-12 : spreading colonial-12
I. Right of mat (13%; 35%). Recombines with mo-5 (5%), spco-11 (43%) (719).
Forms colonies with a downy center and a lacy growing border (719). Hyphal growth is highly branched (2108). Nearly all septae are plugged in peripheral hyphae (2109). Cell-wall peptides are reduced in amount (2249).
spco-13 : spreading colonial-13
VI. Linked to trp-2 (16%), Cen-VI (1/10 asci) (719).
May be allelic with either spco-7 or moe-2. Intercrosses and heterokaryons were unsuccessful with both (719).
spco-14 : spreading colonial-14
II. Linked to arg-5 (7%) (719).
Spreading colonial morphology. A few scattered conidia are produced. Complements da and bal (719).
spco-l5 : spreading colonial-15
III. Linked to spg (l0%), pro-1 (l8%), Cen-III (0/l0 asci). Recombines with col-14, col-16. Complements mo-4, col-14, and col-16 (719).
The original spco-15 mutation has probably been lost. A morphological mutant labeled spco-15 (R1537, FGSC No. 2389) is not linked to III markers (1582).
spe : spermidine
The polyamines spermidine and spermine are synthesized from ornithine in the greater arginine biosynthetic pathway (Fig. 11) via ornithine decarboxylase (EC 4.1.1.17) to putrescine, spermidine synthase (EC 2.5.1.16) to spermidine, and spermine synthase (EC 2.5.1.22) to spermine (Fig. 62). The spe-1 gene encodes the first enzyme. spe-2 encodes S-adenosylmethionine decarboxylase (SAM decarboxylase) (EC 4.1.1.50). This catalyzes the formation of decarboxylated SAM, which reacts with putrescine to form spermidine and methylthioadenosine, a step catalyzed by spermidine synthase, which is encoded by spe-3. In the last step, another molecule of decarboxylated SAM reacts with spermidine to form spermine and methylthioadenosine catalyzed by spermine synthase, the gene for which is as yet uncharacterized.
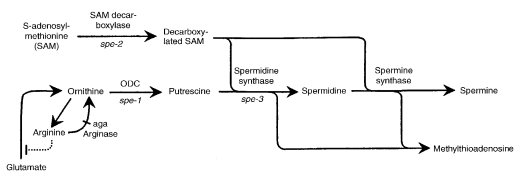
FIGURE 62 The polyamine pathway. The dotted line indicates feedback inhibition of ornithine synthesis by arginine. From ref. (464 ), with permission from the Genetics Society of America.
spe-1 : spermidine-1
VR. Between am (1%) and his-1 (1%) (1626).
Cloned and sequenced: Swissprot DCOR_NEUCR, EMBL/GenBank L16920, M68969, M68970, PIR A42065, EMBL NCODC, NCODCA, NCSPE1A, GenBank NEUODC, NEUODCA, NEUSPE1A; EST NC1F9; Orbach-Sachs clones X3H05, X4E02, G3H04, G4G01, G6H12, G11H12, G17G10, G22F01, G23B02, G24C11.
Structural gene for ornithine decarboxylase (ODC) (EC 4.1.1.17) (611, 2218) (Figs. 11 and 62). Uses putrescine, spermidine, or spermine. Does not suppress pro-4 (1310). Excretes yellow pigment into synthetic cross medium (1546). Meiosis is normal in homozygous crosses (on 5 mM spermidine); these produce mostly white ascospores and a few viable black ascospores (451, 1673). The stability of ODC is regulated by polyamine availability (96, 540). Molecular studies on regulation (1627). Effects on induction by polyamines (805). In contrast to other systems, Neurospora crassa ODC does not appear to be regulated by osmoticum (470). Called put-1.
spe-2 : spermidine-2
VR. Left of am (6%), spe-1 (1626).
Cloned and sequenced: EMBL/GenBank AF151380; Orbach-Sachs clone X2B09 (925).
Specifies S-adenosylmethionine decarboxylase (EC 4.1.1.50) (Fig. 62). Blocked in the conversion of 5-adenosylmethionine (SAM) to decarboxylated SAM (1626). Grows on spermidine, but not on putrescine.
spe-3 : spermidine-3
VR. Right of inl (4%) (1626). Linked to cya-2 (1035).
Cloned and sequenced: EMBL/GenBank AB001598; Orbach-Sachs clone G2212C (1035).
Specifies spermidine synthase (EC 2.5.1.16). Blocked in the conversion of putrescine to spermidine (1626) (Figs. 11 and 62). Cloned independently by suppression of the temperature sensitivity of cpz-2 mycelial growth. Encodes a sequence homologous to spermidine synthase of Saccharomyces, human, and mouse (1035).
spg : sponge
III. Between acr-2 (1%; 11%) and ser-1 (8%). Linked to sc (<1%), thi-4 (0/103) (1582, 1592).
Forms conidiating spreading colonies. Morphologically distinct from sc. Hyphae fuse to form bundles (719). Scanning EM photograph; few major constriction chains are seen (1955). Viability is good. Growth on Vogel’s minimal medium (2163) is less spreading than on crossing medium (1582, 2208). Cell-wall peptides are reduced in amount (2249).
Spiral Growth
Hyphae of the laboratory wild type grow as a loose clockwise spiral in young colonies on the surface of agar (141). The pattern is made more clear if cellophane or another membrane is used to prevent growth into the agar. Strains have been described that show increased (141) or decreased (1902) tightness of coiling or anticlockwise coiling (2256); see coil-1, coil-2, rca-1, rco-1.
sre : siderophore regulation
Unmapped.
Cloned and sequenced: EMBL/GenBank AF087130.
Encodes a GATA-binding protein that controls iron transport by controlling the synthesis of siderophores (2302). SRE and its homologs form a subfamily of GATA factors with two zinc fingers separated by a long stretch of amino acids (2302).
ss : synaptic sequence
IL. Linked to nit-2 (<0.2%) (313).
When alleles ssE, ssS, and ssC are heterozygous (from Emerson, St. Lawrence, and Costa Rica wild types), recombination in nit-2 is reduced 2- to 20-fold, but crossing over in the flanking intervals un-5 to nit-2 or nit-2 to leu-3 is not affected. ss heterozygosity acts multiplicatively with rec-l+ to reduce recombination in nit-2 by l00-fold (313).
ssp-1 : site-specific parvulin-1
Unmapped.
Cloned and sequenced: EMBL/GenBank AJ006023, EMBL NCR6023.
Encodes a homolog of site-specific parvulin (EC 5.2.1.8) (1088), which is active in isomerizing small peptides and in protein folding. Unique among known eukaryotic parvulins in containing a polyglutamine stretch between the N-terminal WW domain and the C-terminal peptidyl-prolyl cis-trans-isomerase (PPIase) domain.
ssu : supersuppressor
Nonsense suppressors (either proved or putative). In a few cases, putative missense mutants also were suppressed (1461). Allele-specific but not locus-specific. Often infertile or only slightly fertile (ssu-3) when used as female parent, forming no perithecia or empty perithecia (1859). Mutants ssu-1, ssu-2, ssu-3, ssu-4, and ssu-5, but not ssu-6, are cold-sensitive in combination with am, showing less than half the wild-type growth rate on minimal medium at 10ºC (1752). Action of the suppressors is nucleus-restricted in heterokaryons (773). Interaction of certain supersuppressors with certain ad-3B alleles produces erratic stop-start growth (772). For tabular summaries of the action of individual ssu mutations on differential test alleles, see refs. (303), (1860), and (1862).
ssu-1 : supersuppressor-1
VIIR. Between met-7 (14%) and su(trp-3td201)-1 (10%), nt (23%) (1859).
Allele WRN33 was selected in ref. (1858) as a suppressor of the amber mutant am17 (1861); tyrosine is inserted in a site where wild type has glutamate (1861). Used to identify suppressible amber mutants in aro(p), trp-1, trp-2, trp-3 (294, 348, 1858), ad-3B (1461), arg-6 (474), nit-2 (1610), and nit-3 (1610). Restores full-length NIT3 to nit-3 amber mutants (1494). Although perhaps the most efficient of known ssu mutants, ssu-1 restores only about 20% of the wild-type amount of normal glutamate dehydrogenase in am17 (1861).
ssu-2 : supersuppressor-2
I. Linked to mat (22%). Probably between Cen-I (7%) and al-2 (26%). Recombines with ssu-3 (1859).
Amber suppressor. Allele WRU35, selected in ref. (1859) as a coincident suppressor of the amber mutants trp-3td140 and am17 (258, 1861). Suppresses the same alleles as ssu-1. Also suppresses certain mutants in trp-1, trp-2, and ad-3B (1860).
ssu-3 : supersuppressor-3
I. Linked to mat (22%). Probably between Cen-I (10%) and al-2 (33%) (1859).
Amber suppressor. Allele WRU118, selected in ref. (1859) as a coincident suppressor of the amber mutants trp-3td140 and am17 (258, 1861). Fails to suppress most amber mutants that are suppressed by ssu-1 and ssu-2 (1860). Probably specifies the insertion of a different amino acid than is inserted by ssu-1 or ssu-4 [T. W. Seale and A. Kinniburgh, cited in ref. (1860)].
ssu-4 : supersuppressor-4
VIIL. Between nic-3 (28%) and met-7 (20%) (1859).
Amber suppressor. Allele WRU18, selected in ref. (1859) as a suppressor of the amber mutants trp-3 (td140) and am (17, 258, 1861). Also suppresses certain mutants in trp-1 and ad-3B (1461, 1860).
ssu-5 : supersuppressor-5
III or IV (303).
Amber suppressor. Allele Y319-45 was selected as suppressor of a nonsense mutant in aro-1(p) (303). Also suppresses the amber mutant trp-3td140 and certain ad-3B alleles, but not am17 (303, 1461, 1862).
ssu-6 : supersuppressor-6
VR. Linked to his-1 (4%) (303).
Amber suppressor. Allele Y319-26 was selected as suppressor of an amber mutant in aro-1(p) (303). Also suppresses certain alleles in trp-3 and his-3, but not am17 (303, 1461, 1862).
ssu-7 : supersuppressor-7
VIL. Between ad-8 (8%) and ylo-1 (14%) (1859).
Amber suppressor. Allele WRU7 was selected in ref. (1859) as a coincident suppressor of the amber mutants trp-3ttd140 and am17 (258, 1861). Also suppresses certain alleles at trp-1, trp-2, and ad-3B. ssu-7 shows the widest spectrum of the known supersupressors (1860). The Y319-37 allele of ssu-8, which is in IR, was erroneously called ssu-7 in some FGSC lists and in ref. (772).
ssu-8 : supersuppressor-8
IR. Linked to al-2 (2%; 8%) (303).
Amber suppressor. Allele Y319-37, selected as a suppressor of an aro(p) amber mutant (303). Also suppresses certain alleles in trp-3, his-3 (303), and ad-3B (1461). Allele Y319-37 (FGSC No. 1749), also called 54-su37, was thought possibly to be an allele of ssu-2 and was listed as ssu-2 (?) before being designated ssu-8 (303). It was erroneously called ssu-7 in various FGSC lists and in ref. (772) and was erroneously stated to be in linkage group VI (the location of the real ssu-7).
ssu-9 : supersuppressor-9
Unmapped. Locus distinct from other ssu genes (1860).
Amber suppressor. Allele WRU98 was selected (1860) as a coincident suppressor of the amber mutants trp-3td140 and am17 (258, 1861). Also suppresses certain alleles at trp-1 and trp-2 (1860).
ssu-10 : supersuppressor-10
Unmapped. Locus distinct from other ssu genes (1860).
Amber suppressor. Allele RWU121 was selected (1860) as a coincident suppressor of the amber mutants trp-3td140 and am17 (258, 1861). Also suppresses certain alleles at trp-1, trp-2, and ad-3B (1860).
ssu(WRU79) : supersuppressor (WRU79)
Unmapped. Locus distinct from other ssu genes (1860).
Amber suppressor. Selected (1860) as a coincident suppressor of the amber mutants trp-3td140 and am17 (258, 1861). Also suppresses certain alleles at trp-1, trp-2, and ad-3B. Spectrum resembles ssu-1 (1860).
st : sticky
IR. Between ad-3B (5%) and thi-1 (14%) (1548).
Mycelia adhere to an inoculating needle. Subtly different from wild-type in gross morphology. Exudate is sometimes present (1548). Grows poorly from conidia inoculated to minimal agar medium with ballooning of hyphal tips, which is not alleviated by mannose (1546).
Sterile Mutants
The unqualified term "sterile" has been used in a variety of ways for different situations, such as where no protoperithecia are formed, where perithecial development is blocked before ascospore formation, or where perithecia develop fully but the ascospores that are produced are inviable. To avoid ambiguity, the type and extent of sterility should be specified (1577). The term "barren" has been proposed for crosses that form perithecia but produce few or no ascospores (1685). See ff, fmf, fs, mb, pp.
su : suppressor
The symbol of the gene that is suppressed follows su in parentheses. The mutant suppressor allele is symbolized su(. .) and the wild-type nonsuppressor, su(. .)+. ropy mutations can be obtained as suppressors of cot-1 (247, 1634). crisp mutations can be obtained as suppressors of mcb (247). For suppressors of vegetative incompatibility, see tol, su(het). arg-12S acts as a suppressor of pyr-3.
su(arg-1)-1 : suppressor-1 of arg-1
Unmapped. Not linked to arg-1.
Restores 23-36% of wild-type L-citrulline:L-aspartate ligase activity to arg-1 allele 46004. Called arg-1R26, arg-1R3, s-26, s-3 (122).
su(bal) : suppressor of bal
I. Linked to mat (13%) (1853).
The linear growth rate of bal is doubled and the Km of G6PD is decreased by the suppressor. Photographs are given of bal and bal; su(bal) (1853). Called su-B.
su(col-2) : suppressor of col-2
IL. Tightly linked to mat (0/837) (1853).
Increases the linear growth rate of col-2 10-fold and influences the electrofocusing pattern of G6PD both in col-2 and in col-2+. The morphology of su(col-2); col-2+ is wild type. Photographs are given of col-2 and col-2; su(col-2) (1853). Called su-C.
su(cot-1) : suppressor of cot-1
A series of mutants selected as suppressors of the colonial growth of cot-1 were named gulliver (gul). ropy mutations can be selected as suppressors of cot-1, and existing ropy mutants act as cot-1 suppressors.
su(cr-1) : suppressor of cr-1
See hah. Suppressors of the colonial growth of cr-1 have also been reported in ref. (1020), with alterations in cAMP level, trehalase activity, conidiation, and pigmentation .
su(het) : suppressor of heterokaryon (vegetative) incompatibility
The first described het suppressor, tol (1466), acts only on the vegetative incompatibility function of the mating-type idiomorphs (1175). Mutations since have been reported that are interpreted to suppress vegetative incompatibility at other het loci (51). Some of these mutations are reported to affect only single loci, others to prevent incompatibility reactions even when strains differ at several het loci. One mutation, thought at first to be a suppressor of het-d, was reinterpreted as a new het locus called het-12 (1492). Characterization and mapping of an extragenic het-suppressor mutation, su(het-d), has been reported (1345, 1346). Interpretation of the results reported in ref. (51) has been complicated by the subsequent discovery that the strains used may differ at another locus or other loci affecting heterokaryon formation (980, 2223).
su(het-d) : suppressor of het-d
Appears to suppress het-d mediated vegetative incompatibility. The difference from wild type is less marked in young cultures (1345, 1346).
su(ile-1) : suppressor of ile-1
VR. Right of met-3 (4%) (1055).
Suppresses the requirement of ile-1 (1055).
su(inl) : suppressor of inositol
Unmapped. Not linked to inl (735).
An allele-specific partial suppressor that enables inl allele 37401 to grow suboptimally on minimal medium (735). Does not suppress seven other inl alleles, including 89601, that is partially suppressed by opi-1.
su(mcb) : suppressor of mcb
See cr-1. cr-1 mutations can be selected as suppressors of the apolar growth of mcb (247).
su(met-2) : suppressor of met-2
Unmapped. Not tested for allelism with su(met-7)-1 or su(met-7)-2.
Isolated as a suppressor of met-2H98 (2012). Suppresses leaky mutants blocked between cysteine and homocysteine. It is probably the suppressor used in ref. (2211) to show that the suppressed strain differs from wild type in being able to incorporate sulfur from S-methylcysteine into cysteine regardless of the sulfate concentration. The suppressor does not have significantly increased acetylhomoserine sulfhydrylase (1045).
su(met-7)-1 : suppressor-1 of met-7
IR. Linked to al-2 (1%) (727).
Selected as reversions of met-7 (4894) in ref. (727). All spontaneous reversions were mutations at this locus. Also suppresses met-2 (H98). Not tested for suppression of other alleles or loci. Suppressed strains attain wild-type growth on minimal medium after initial retardation, which is alleviated by methionine (727). This is apparently the suppressor used in ref. (649) to show that the suppressor restores cystathionase I and II activities to 4894 and H98, respectively. Called S-1, FGSC 39.
su(met-7)-2 : suppressor-2 of met-7
Unmapped.
Suppresses met-7 (4894). Not tested for suppression of other mutants. Selected as a revertant of met-7 (4894) in ref. (727). Recovered less frequently than su(met-7)-1 and only after irradiation. Called S-2.
su([mi-1])-1 : suppressor-1 of[mi-1]
IIIR. Linked to ad-4 (14%) (1084, 1085).
Restores normal growth rate and alleviates deficient cyanide-sensitive respiration of [mi-1] and other group I mitochondrial mutants. No effect on group II or III mitochondrial mutants (168, 400, 778, 1085). Mitochondrial ribosomal subunit ratios and cytochrome spectrum (400). Called sup-1.
su([mi-1])-3 : suppressor-3 of [mi-1]
II. Linked to fl (31%). Not allelic with su([mi-1])-4 or su([mi-1])-10; 20% and 25% unsuppressed progeny from intercrosses (1084, 1085).
Restores normal growth rate and alleviates the deficient cyanide-sensitive respiration of [mi-1] ([poky]) and another group I cytoplasmic mutant. Does not affect group II mitochondrial mutant [mi-3] (168, 400, 1085). Mitochondrial ribosomal subunit ratios and cytochrome spectrum (400, 1085).
su([mi-1])-4 : suppressor-4 of [mi-1]
II. Linked to fl (22%), arg-5 (40%). Not allelic with su([mi-1])-3 or su([mi-1])-10; 20% and 25% unsuppressed progeny from intercrosses (1084, 1085).
Restores normal growth rate and alleviates the deficient cyanide-sensitive respiration of [mi-1] ([poky]) and another group I cytoplasmic mutant. Does not affect group II cytoplasmic mutant [mi-3] (168, 400, 1085). Mitochondrial ribosomal subunit ratios and cytochrome spectrum (400). Called sup-4 (1085).
su([mi-1])-5 : suppressor-5 of [mi-1]
VIIL. Left of nic-3 (23%) (1084, 1085). Possibly allelic with cyt-7 (170).
Restores normal growth rate and alleviates the deficient cyanide-sensitive respiration of [mi-1] ([poky]) and another group I mitochondrial mutant. Does not affect group II cytoplasmic mutant [mi-3] (168, 400, 1085). Mitochondrial ribosome and cytochrome spectra (400). Affects mitochondrial large-subunit assembly. With wild-type mitochondria, causes cold sensitivity (399). Called sup-5 (1085), suI-5 (399).
su([mi-1])-10 : suppressor-10 of [mi-1]
II. Linked to arg-5 (29%), fl (24%) (1084, 1085). Not allelic with su([mi-1])-3 or su([mi-1])-4; 25% unsuppressed progeny from intercrosses (1085).
Restores normal growth rate and alleviates deficient cyanide-sensitive respiration of [mi-1] ([poky]) and all other group I cytoplasmic mutants. Does not affect group II cytoplasmic mutant [mi-3] (168, 400, 1085). Cytochrome spectrum (400). Called sup-10 (1085).
su([mi-1])-14 : suppressor-14 of [mi-1]
IV. Linked to arg-2 (14%) (1084, 1085).
Restores normal growth rate and alleviates the deficient cyanide-sensitive respiration of [mi-1] ([poky]) and all other group I cytoplasmic mutants. Has no effect on group II cytoplasmic mutant [mi-3] (168, 400, 1085). Cytochrome spectrum (400). Called sup-14 (1085).
su([mi-1])-f : suppressor-f of [mi-1]
VR. Left of inl (10%) (1915).
Restores normal growth rate. Alleviates the deficient cyanide-sensitive respiration of [mi-1] ([poky]) (1370) and all other group I mitochondrial mutants, but not of group II or group III mutants (171, 739) (Appendix 4). su([mi-1])-f differs from other known [mi-1] suppressors in not restoring SHAM insensitivity (1085). Cytochrome spectrum (168). Used for studying oscillations in membrane potential (763). Called f.
su([mi-3])-1 : suppressor-1 of [mi-3]
IR. Between nit-1 (17%), un-7 (£1%) and al-2 (1%; 4%) (739).
Restores normal growth rate and cytochrome spectrum to [mi-3], a group II mitochondrial mutant (172, 739). Does not suppress the mutant phenotype of group I or group III mitochondrial mutants (171).
su(mtr)-1 : suppressor-1 of mtr
IR. Right of his-2 (2%) (1991).
Selected in trp-1; mtr by increased uptake of tryptophan (1991) or as a his+ revertant of his-2; mtr on excess arginine (228). Still resistant to 4-methyltryptophan, but now sensitive to p-fluorophenylalanine (1991). The effect of su(mtr)-1 on mtr is locus-specific. su(mtr)-1 appears to have a changed regulation for amino acid transport system II (1520). May be allelic with lysR (1082).
su(mtr26) : suppressor of mtr26
VIR. Linked to pan-2, trp-2. Right of Cen-VI (227).
Allele-specific partial suppressor of the poor growth of trp-1; mtr26 on low levels of tryptophan. [mtr2 6 is a putative frameshift (227).]
su(pan-2Y153M66) : suppressor of pan-2Y153M66
Unmapped.
Allele-specific. Not effective on three other pan-2 alleles or on four supersuppressible alleles at trp-3 and am-1 (357).
su([poky]) : suppressor of [poky]
See su([mi-1]).
su(pro-3) : suppressor of pro-3
Allelic with arg-6.
su(suc)-1 : suppressor-1 of suc
Unmapped. Unlinked to suc (1119).
Isolated as a revertant of suc KG163, which is associated with a putative inversion in IL. Grows on acetate-free minimal medium, but the growth is enhanced by acetate supplementation. Suppresses other suc mutants but not mutations at six ace loci (63). Growth is yeastlike in acetate-free, vigorously agitated, liquid culture at 37ºC. Called R2, 163R2 (63, 1119).
su(suc)-2 : suppressor-2 of suc
I. Linked to suc (3%) (1119).
Isolated as a revertant of suc(KG163), which is associated with a putative inversion in IL. Grows on acetate-free minimal medium, but the growth is enhanced by acetate supplementation. Forms spheroplasts and short chains of yeastlike cells when grown at 34ºC in acetate-free, vigorously agitated, liquid culture. Called 163R4 (63, 1119).
su(trp-3) : suppressor of trp-3
Early work with trp-3 suppressors is reviewed in ref. (2264). trp-3 mutants were originally called td, and alleles were designated td2, td3, etc. Most suppressors of trp-3 were assigned numbers corresponding to those of the allele in which they originally were discovered. With td2, the td number also was retained as a suppressor locus number. Thus, the first suppressor discovered in td2 is symbolized su(trp-3td2)-2. The locus number 2 does not imply that there was a previously discovered suppressor of the td2 mutation. A second suppressor gene discovered in td2 was numbered 2a and is now symbolized su(trp-3td2)-2a. In contrast, when different suppressors of the td201 mutation were found, they were treated conventionally and given consecutive locus numbers beginning with 1.
su(trp-3td2)-2 : suppressor-2 of trp-3td2
III. Linked to leu-1 (22%) (1126).
Allele-specific. Suppresses allele td2 but does not suppress td6, td71, or any other allele from td1 through td34 (1125, 2266). This and the other listed suppressors of td2 were not tested on later trp-3 isolates that fall in the same complementation group as td2. Although suppressed mutants grow on minimal medium, growth is stimulated by the addition of tryptophan. Suppressed trp-3; su colonies are morphologically distinguishable from wild type. Tryptophan synthetase is formed at levels below wild type. Originated in trp-3td2 (originally numbered S1952) (2262, 2266). Called su-2 (729), su2 (2266).
su(trp-3td2)-2a : suppressor-2a of trp-3td2
I. Linked to al-2 (15%) (1126).
Isolated in ref. (2263) as one of four suppressors of trp-3td2, called su2a, su2b, su2c, su2d. Of the four, only su2a was mapped. All are nonallelic with su2 and with each other, with the possible exception of su2b and su2c (2263). None was tested for allelism with su(trp-3td6), which also suppresses trp-3td2. The a that follows the locus number does not refer to mating type. Called su-2a (1126).
su(trp-3td3): suppressor of trp-3td3
Unmapped.
Allele-specific. Suppresses trp-3 alleles td3, td24, and td71, but not any other mutants from tdl through td34 (1125, 2266). Originated in trp-3td3. Probably allelic with su(trp-3)td24. Called su3 (2266).
su(trp-3td6) : suppressor of trp-3td6
Unmapped. At a different locus from su(trp-3td2)-2 and su(trp-3td3).
Allele-specific. Suppresses trp-3 alleles td6 and td2 but does not suppress any other allele from td1 through td34 (1125, 2266). Although suppressed mutants grow on minimal medium, growth is stimulated by the addition of tryptophan and is morphologically distinguishable from wild type. Tryptophan synthetase is formed at levels below wild type. Originated in trp-3td6. Called su6 (2266).
su(trp-3td201)-1 : suppressor-1 of trp-3td201
VIIR. Between met-7 (18%), ssu-1 (10%; 13%) and arg-10 (7%) (1859, 2284).
Allele-specific. Suppresses missense allele td201, but not other alleles: tdl, td6, td7, td16, td37R, td71, td138R, td141 (2284), td2, td3, or td24 (1974). A suppressed mutant has a low level of tryptophan synthetase activity, allowing slow growth on minimal medium. Enzyme activity is due to a protein physically like the wild-type enzyme (2284, 2285). Suppressor alone, without td201, grows slightly slower than wild type (1974). The suppressor is effective when in another nucleus from the suppressible td201 in a forced heterokaryon between noncomplementing alleles (trp-3td16; su + trp-3td201; su+) (1974). Called Su-1 or Su-1td201 (1859), su-YS (1974), su1(201) (2283).
su(trp-3td201)-2 : suppressor-2 of trp-3td201
VII. Linked to su(trp-3td201)-1, probably to the left (1974).
Allele-specific. Suppresses missense allele td201, but does not suppress alleles td1, td2, td3, td16, td24, or td71 (1974). A suppressed mutant has a low level of tryptophan synthetase activity, allowing slow growth on minimal medium. Enzyme activity is due to a protein physically distinguishable from wild-type tryptophan synthetase, unlike suppressors 1 and 3 (1658). Suppressor alone, without td201, grows slightly more slowly than wild type (1974). The suppressor is effective when in another nucleus from td201 in a forced heterokaryon between noncomplementing alleles (trp-316; su + trp-3td201; su+) (1974). Called su-R (1974), su-2 (1658).
su(trp-3td201)-3 : suppressor-3 of trp-3td201
Unmapped. Not linked to su(trp-3td201)-1 or other VII markers.
Allele-specific. Suppresses missense allele td201, but not alleles td1, td6, td71, or td141. The one su(trp-3td201)-3 allele tested gave less powerful suppression of td201 than did the known alleles of su(trp-3td201)-1. The suppressed mutant has enough tryptophan synthetase activity to allow slow growth on minimal. The restored enzyme activity is due to a protein physically like wild type. Called su3, su3(201) (2283).
su(trp-5) : suppressor of trp-5
VIL. Closely linked to (or allelic with) aro-6 (1055).
Possibly a feedback-negative mutant of aro-6, which specifies DAHP synthase (Tyr) (1055).
su(ure-19) : suppressor of ure-19
Unmapped. Not linked to ure-1 or ure-2.
Suppressor of ure-1 allele 9. Does not suppress ure-247. Only ure-19 strains from a certain lineage are suppressed, suggesting that suppression requires a cosuppressor closely linked to ure-19 (255).
suc : succinate
IL. Between acr-3 (2%) and In(H4250)L, phe-1 (1%) (1122, 1592)
Uses acetate, succinate, or any of numerous related compounds (1187). Most, but not all, strains grow better on acetate than on succinate (1119). Lacks pyruvate carboxylase activity (EC 4.1.1.31) (140, 1122, 2009). Numerous alleles are CO2-remediable (230). Leaky, but less so on higher ammonium (2008). A special medium has been devised for selective plating [K. Munkres, unpublished; see p. 248 in ref. (1592)]. Most ascospores are poorly pigmented in homozygous suc ´ suc crosses. Allele KG163 shows greatly reduced recombination in the region between leu-4 and suc, suggesting inversion heterozygosity (1119). suc would be named ace-6 except for priority of nomenclature (1122).
Sulfur Metabolism
Reviewed in refs. (1283) and (1287). See Figs. 24 and 45.
sup : suppressor
Symbol changed to su.
sw : snow white
Changed to per-1.
Return to the 2000 Neurospora compendium main page