The single letter italicized has been used as a gene symbol (T"> T : Tyrosinase
IR. Between ad-3A (18%) nic-2 (12%) and al-2, al-1 (12%) (351, 897).
Cloned and sequenced: Swissprot TYRO_NEUCR, EMBL/GenBank M32843, M33271, PIR A00511, A34460, YRNC, EMBL NCTYRA, NCTYRB, GenBank NEUTYRA, NEUTYRB; pSV50 clones 30:2D, 32:9A.
Tyrosinase structural gene (EC 1.14.18.1) (Fig. 13). Prototrophic. Multiple alleles from different original strains were distinguished electrophoretically and by thermolability (896, 897, 899-901, 1748). The primary structure (407 amino acids) was determined by amino acid sequencing the product of allele TL (1163). The polypeptide contains a cysteine and a histidine linked by a thioether bridge (1165, 1167, 1169). Precursor contains a carboxy-terminal extension (1115). Two forms of enzyme were demonstrated in a heterokaryon (897). Scored by color reaction with DL-DOPA as substrate (899, 901). For regulation, see refs. (623), (898), and (899). Tyrosinase synthesis is induced by a variety of compounds, including cycloheximide and amino acid analogs. Sulfhydryl compounds can also affect tyrosinase expression (1646). The addition of copper before cycloheximide induction increases the yield of enzyme (936). Extensive biochemical and biophysical analyses of the enzyme (149, 150, 152, 1166, 1170, 1613, 1749, 2103, 2244); enzyme crystallized (692). Used as a reporter gene (1090, 1114). Tyrosinase regulatory genes are symbolized ty.
t : thermophobic
Probably a scot allele. See scot.
T(. . .) : Translocation
A rearrangement in which segments of two or more chromosomes are interchanged or a segment has been moved from one chromosome to another. Symbolized in full by T followed by the involved linkage group or groups in parentheses and an identification number, all in italics. For example, T(IR;IVR)NM172, T(IL®IIR)39311. In the present text and in maps, the symbol is abbreviated with the linkage groups omitted and the identification number in parentheses. For example, T(NM172), T(39311). Major types of translocations are reciprocal, in which two terminal segments are interchanged; insertional, in which a segment is removed from one chromosome and inserted in another and quasiterminal, in which a distal segment is translocated to the tip region of another chromosome, distal to any essential gene. Insertional and quasiterminal rearrangements are useful for determining dominance and for mapping by duplication coverage (1566). See ref. (1578) for information on individual translocations.
T(OY329) : Translocation (VIR®IIIR)OY329
Probably a quasiterminal translocation. [Misdiagnosed as insertional in ref. (1578)]. A segment of VIR that includes trp-2, ws-1, and col-18 (but not del) apparently is translocated to Tip-IIIR.
ta : tufted aerial
IL. Between un-l6 (2%) and acr-3 (<3%) (1582, 1592).
Growth is colonial and rapid-spreading. Morphology varies with growth conditions (1592). Conidiation is best at 34ºC. There may be a maternal effect, with ta+ resembling ta in initial cultures from ascospores (1476).
Tad : Transposable element Adiopodoume
Cloned and sequenced: GenBank/EMBL L26562, L26563, GenBank NEUGLPRCA, NEUGLPRCB.
A LINE-like retrotransposon present as an active element in a strain from the Ivory Coast and discovered when the am gene was inactivated by insertion of Tad (1065). Active Tad elements can be introduced into laboratory strains by crosses, heterokaryons, or transformation. In heterokaryons, an active Tad is transferred readily to nuclei that previously lacked it (1932); if Tad in the donor nucleus contained an intron, the intron is spliced out prior to transfer (1061). When a strain that previously lacked an active Tad acquires it, the element remains active during vegetative growth and propagation. It is inactivated by RIP when it goes through a cross, however. Laboratory strains and strains representing seven heterothallic and homothallic Neurospora species contain inactive Tad-like sequences, and these appear to have undergone RIP (1064).
tba-1 : tubulin alpha -1
Unmapped.
Cloned and sequenced: Swissprot TBA1_NEUCR, EMBL/GenBank X79403, GenBank NCATUBA, PIR S45050.
Encodes the α-a chain of tubulin. Developmentally regulated; RNase protection indicates that the mRNA is not present during the first 30 min of conidial germination (1375).
tba-2 : tubulin alpha-2
Unmapped.
Cloned and sequenced: Swissprot TBA2_NEUCR, EMBL/GenBank X79404, PIR S45051, GenBank NCATUBB; EST NP6G3
Encodes the α-b chain of tubulin. Transcript is present throughout the vegetative cycle. Shows only 65% amino acid sequence homology with the product of tba-1 (1375).
tbg : tubulin gamma
Unmapped.
Cloned and sequenced: Swissprot TBG_NEUCR, EMBL/GenBank X97753, GenBank NCGTUB.
Encodes g-tubulin (864).
Tcen : Centromeric transposable element
VII. In Cen-VII and other centromere regions (275).
Sequenced (275).
A full-length Copia-like transposable element found in Cen-VII, where it is clustered with degenerate fragments of Tgl1, Tg12, and Tad (275).
td : tryptophan desmolase
Changed to trp-3, which encodes tryptophan synthetase (the renamed enzyme).
tef-1 : translation elongation factor-1
IL. Linked to leu-4, mat, arg-3 (0/18) (1447).
Cloned and sequenced: Swissprot EF1A_NEUCR, EMBL/GenBank D45837, GenBank NEUEF1A; EST NM6E1; Orbach-Sachs clones X9B04, X9E03, X25C03, X25G03.
Structural gene for translation elongation factor-1α. The mutant isolated by RIP has a slow-growth phenotype (944). Cycloheximide inducible. Expression at the level of mRNA appears constitutive in the lah-1 single mutant, but not in a lah-1; cpc-1 double mutant (833).
Tel : telomere
Neurospora telomeres consist of tandem repeats of TTAGGG (identical to the telomere sequence in humans). The prototype in VR was identified by walking from the distal marker his-6 (1809). The oligonucleotide (TTAGGG)4 was then used as a hybridization probe to show that the repeated sequence is found only at chromosome ends and to identify restriction fragments containing telomeres and subtelomeric regions from other chromosome tips (1810).
Tel-IR : Telomere-IR
IR. Linked to arg-13 (6T, 2NPD/18 asci) (1447).
Cloned (1447).
Telomere-derived restriction fragment M7 (1810).
Tel-IIL : Telomere-IIL
IIL. Linked to hsp70 (2T/18 asci), Fsr-52, vph-1 (14T + 1 double crossover/18 asci) (1447, 1810).
Telomere-derived restriction fragments 014, M5 (1810).
Tel-IIR : Telomere-IIR
IIR. Right of leu-6 (1T, 1NPD/16 asci) (1447, 1810).
Telomere-derived restriction fragment 05 (1810).
Tel-IIIR : Telomere-IIIR
IIIR. Linked to cat-3 (5T/18 asci), Fsr-45 (3T, 1NPD/18 asci). Fsr-45 in turn is linked to ro-2 (3/18), trp-1 (6/18) (1447, 1810).
Telomere-derived restriction fragments 07, M13 (1810).
Tel-IVL : Telomere-IVL
IVL. Linked to T(AR33) (1/20) (1334, 1811). Left of sdv-1, -2, -3 (12T, 0NPD/18 asci) (1447).
Telomere-derived restriction fragment 010 (1810).
Tel-IVR : Telomere-IVR
IVR. Linked to nuo21 (1T/18 asci) (13T, 2NPD/18 asci with trp-4) (1447).
Telomere-derived restriction fragment M3 (1810).
Tel-VR : Telomere-VR
VR. Linked to his-6 (30 kb) (1447, 1809-1811).
Cloned and sequenced: EMBL/GenBank M37064, GenBank NEUTELVRA.
Telomere-derived restriction fragment M14 (1810). Cloned by walking from his-6 (1809). The first Neurospora telomere to be cloned.
Tel-VIL : Telomere-VIL
VIL. Linked to cax (3T/18 asci) (1447). Assignment of Tel-VIL (and cax) to VIL depends on the probable linkage of Tel-VIL to tf2d (7T, 1NPD/16 asci) (1447). The tf2d sequence is included in the duplicated segment generated in crosses of T(VIL®IR)T39M777 ´ Normal (1322).
Telomere-derived restriction fragments 011, M10 (1322, 1810).
Tel-VIR : Telomere-VIR
VIR. Linked to the terminal VIR breakpoint of T(UK-T12) (0T/9 asci; 0/13) (64) and to nuo21.3 (0T/18 asci), con-11a (9T/18 asci) (1447).
Telomere-derived restriction fragment M11 (1810).
Tel-VIIL : Telomere-VIIL
VIIL. Linked to nic-3, pho-4 (10T, 1NPD/18 asci) (1447).
Telomere-derived restriction fragments 09, M9 (1810).
tet : tetrazolium
IL. Linked to acr-3 (2%), ad-3B (l%). Right of mat (7% or 8%) (739).
Detected as a difference between A and a laboratory strains in the ability to reduce tetrazolium dye. Wild-type 74-OR23-1A colonies fail to reduce dye and thus remain white, whereas 74-OR8-1a colonies reduce dye to red (739). Called Tet-R, Tet-W (“red,” “white”). See ref. (739) for tests on other wild types. The same gene is perhaps responsible for a mat-linked difference in resistance to 2,3,5-triphenyltetrazolium chloride (TTC), although linkages appear different. [TTC resistance maps left of mei-3, probably between mat and arg-1 (707).] TTC-resistant strains include 74-OR23-1A, Em A FGSC No. 691, Em A 5256, Lindegren 1A, Lindegren 25a, and all RL wild types tested. TTC-sensitive strains include 74-OR8-1a, Em a FGSC No. 692, and Em a 5297 (2180, 2181). Strain ORSa, derived by backcrossing to 74-OR23-1A, is resistant (1422). cya, cyb, and cyt mutants all fail to reduce TTC, and this is used as a test in the initial identification of such mutants. cya-l is located near mat (170); its relationship to tet is not known.
tf2d : transcription factor IID
VIL. Left of T(T39M777) (included in duplications from T ´ Normal) (1322). Probably linked to cax (6T, 1NPD/18 asci), Tel-VIL (7T, 1NPD/18 asci) (1447).
Specifies TATA box binding transcription factor TfIID (2134).
Tgl1 : Gypsy-like-1 (Transposable Element)
Cloned and sequenced (275).
A degenerate Gypsy-like retrotransposon. Found in Cen-VII as a component of nested clusters containing this and other AT-rich immobile transposable elements with earmarks of having been subjected to RIP. Interrupted by different defective fragments of Tad (275).
Tgl2 : Gypsy-like-2 (Transposable Element)
Cloned and sequenced (275).
A degenerate retrotransposon identified as Gypsy-like by sequence similarity to Ty3-2 of Saccharomyces. Found in Cen-VII as a component of nested clusters containing this and other AT-rich immobile transposable elements with earmarks of having been subjected to RIP (275).
thi : thiamine
Thiamine biosynthesis in fungi involves the condensation of pyruvate and glyceraldehyde-3P to form the thiazole ring and then coupling of this thiazole with (hydroxymethyl)pyrimidine diphosphate by a single-gene-encoded thiamine biosynthetic bifunctional enzyme containing activities for thiamine phosphate pyrophosphorylase (EC 2.5.1.3) (TMP-PPase) and (hydroxyethyl)thiazole kinase (EC 2.7.1.50) [4-methyl-5-(β- hydroxyethyl)thiazole kinase] (THZ kinase), the last also being responsible for a salvage synthetic pathway (2062).
thi-1 : thiamine-1
IR. Between cys-9 (13%), T(4540)R and T(NM103), met-6 (7%; 14%) (1414, 1578, 1592, 2123).
Uses thiamine or its precursors, pyrimidine + thiazole (2060). Adaptation to growth on minimal medium occurs after a lag. Growth tests should therefore be scored early. Adaptation is not carried over via ascospores, conidia, or small mycelial fragments. Adaptive growth is paralleled by the attainment of wild-type thiamine pyrophosphate and carboxylase levels. The function apparently concerns utilization of intact thiamine rather than its biosynthesis (584, 585). T(17084) is inseparable from thi-1 (1578).
thi-2 : thiamine-2
IIIR. Between his-7 (1% or 2%) and ad-2 (1%; 3%) (426, 1587).
Requires thiamine. Cannot use pyrimidine + thiazole (2060); therefore, blocked beyond the early condensation step. Does not undergo growth adaptation on minimal medium (584).
thi-3 : thiamine-3
VIIL. Between nic-3 (9%; 18%), lacc (7%) and T(T54M50), ace-8 (1%) (1120, 1578, 1580, 1585, 1592, 2296). Linked to csp-2 (<1%) (1582).
Uses thiamine or thiazole (2060). Does not undergo growth adaptation on minimal medium. Growth on minimal medium is leaky at first but becomes tight with the exhaustion of endogenous thiazole (584), so that scoring is best done late.
thi-4 : thiamine-4
IIIR. Between Cen-III (1T/18 asci) and ser-1, pro-1 (3%; 1T/18 asci), ace-2 (4%; 9%). Linked to acr-2, spg, sc (<1%) (449, 1447, 1585).
Cloned and sequenced: EMBL/GenBank D45894; Orbach-Sachs clone X14F01
Requires thiamine (583). Believed to be involved in the condensation of pyrimidine and thiazole precursors (28). Analog of Fusarium stress-inducible thiazole biosynthetic enzyme. Very leaky (583). A probable allele called thi-lo greatly increases the thiamine requirement of thi-1 and decreases the ability to synthesize thiamine from pyrimidine and thiazole. thi-lo has no detectable nutritional requirement in the absence of thi-1. No recombination, thi-lo ´ thi-4 (0/55 scorable progeny) (583).
thi-5 : thiamine-5
IVR. Linked to pan-1 (1%) (1585).
Uses thiamine (907). Probably can also use 2-methyl-4-amino-5-(aminomethyl)pyrimidine (1557).
thi-6 : thiamine-6
VIIR. Immediately adjacent to ras-3 (300 bp), near un-10 (1267).
Cloned and sequenced (1267).
Homologous to genes, in yeast and elsewhere, that specify the thiazole precursor of thiamine. The mutant phenotype has not been determined (1267).
thi-lo : thiamine-low
A thi-4 allele.
thr-1 : threonine-1
Changed to ile-1. Because this locus specifies threonine dehydratase, the original name is inappropriate (1056, 1059) (Fig. 45). Called thre-1 (463).
thr-2 : threonine-2
IIL. Between ro-3 (6%; 25%), T(NM149) and T(AR179), bal, arg-5 (3%; 18%). Adjoins thr-3 (<0.1%) (1580, 1582, 1585).
Lacks threonine synthetase (EC 4.2.99.2) (654). Requires threonine. Cannot use other amino acids (2064) (Fig. 45). Strongly inhibited by methionine (606, 2064). Known mutants are not heat-sensitive, unlike all known thr-3 alleles.
thr-3 : threonine-3
IIL. Linked to thr-2 (<0.1%) (1585).
Requires threonine. Also responds slightly to α-aminobutyric acid or isoleucine. Known alleles are heat- sensitive (25 vs 34ºC), tight at 34ºC. Not inhibited by methionine (606, 2064).
ti : tiny
IL. Between arg-3 (1%) and T(39311)R (1582).
Heat-sensitive. Spreading colonial morphology at 25ºC or below, more restricted at 30ºC, and no growth at 34ºC (1548, 1582). Can be scored microscopically after ascospore germination. Cell-wall peptides are reduced in amount (2249).
tim : translocase of inner mitochondrial membrane
Translocase complex of the inner mitochondrial membrane. Reviewed in ref. (1611) (Fig. 64). Genes that specify components include tim8, tim17, and hsp70 (1194). See also tom.
tim8 : translocase of inner mitochondrial membrane 8
Unmapped.
Cloned and sequenced: EMBL/GenBank AF142423.
Encodes a small zinc finger protein involved in mitochondrial carrier import (128).
tim17 : translocase of inner mitochondrial membrane 17
VI. Mapped by RFLP (2046).
Cloned and sequenced (2046).
Specifies the 17-kDa protein subunit of TIM (2046). See Fig. 64.
Tip : linkage group tip
Eight of the 14 tips have been mapped genetically by determining the breakpoints of quasiterminal chromosome rearrangements. A breakpoint is known to be genetically terminal when a class of meiotic segregants is recovered that would necessarily be deficient for any essential gene distal to the breakpoint. Although quasiterminal rearrangements behave genetically as terminal, they are thought to be reciprocal translocations in which only the telomere is translocated. See refs. (1566) and (1578). VL was the first tip to be mapped, using the heteromorphic nucleolus satellite as a terminal cytological marker. Of the six tips that are not involved in quasiterminal rearrangements, all but IIIL and VIIR have been mapped by recombination using RFLPs in cloned telomere regions. See Tel.
Tip-IL : Left tip of linkage group I
IL. Marked by quasiterminal translocation T(5936), which is linked to ro-10 (0/38), fr (11%) (1580, 1582).
Tip-IR : Right tip of linkage group I
IR. Marked by quasiterminal rearrangements In(NM176), In(H4250), and T(T39M777), which are closely linked to un-18, het-5, and R (279, 978, 1578, 2129).
Tel(IR) is linked to arg-13 (1447).
Tip-IIL : Left tip of linkage group II
IIL. Left of T(AR30), which is left of pi (28%) (1173).
Tel-IIL is linked to hsp70 and other RFLP markers in IIL (1447, 1810).
Tip-IIR : Right tip of linkage group II
IIR. Right of un-15, rip-1.
Tel-IIR is linked to leu-6 and other RFLP markers in IIR (1447, 1810).
Tip-IIIL : Left tip of linkage group III
IIIL. Left of r(Sk-2)-1, cum, acr-7.
Tip-IIIR : Right tip of linkage group III
IIIR. Marked by quasiterminal translocations T(DBL9), T(OY320) (1578), which are right of dow (3/30; 2/18), erg-3.
Tel-IIIR is linked to IIIR RFLP markers (1447, 1810).
Tip-IVL : Left tip of linkage group IV
IVL. Marked by quasiterminal translocation T(AR33), which is linked to cys-10 (0/221), left of acon-3 (<1%), uvs-3 (1447, 1578, 1811).
Cloned (1810).
Tel-IVL is linked to IVL RFLP markers (1447, 1810).
Tip-IVR : Right tip of linkage group IV
IVR. Marked by quasiterminal translocations T(AR209), T(T54M50), T(OY337), and T(ALS179), which are right of pmb, uvs-2 (2-6%), cys-4 (1578).
Tel-IVR is linked to IVR RFLP markers (1447, 1810).
Tip-VL : Left tip of linkage group V
VL. Marked by sat, NOR and by quasiterminal rearrangements T(NM169d), T(ALS176), T(ALS182), T(OY321), T(AR190), and In(UK-2y)L, which are left of dgr-1, T(AR30), caf-1 (>11%), and lys-1 (20%; 35%) (118, 1578, 1601).
Tip-VR : Right tip of linkage group V
VR. Marked by quasiterminal translocation T(NM149) (1578, 1809).
Cloned and sequenced: GenBank M37064.
Tel-VR is linked to his-6. RFLP analysis (1810). A copy of the retrotransposon Pogo adjoins Tel-VR in strains of OR background (1447, 1809, 1811).
Tip-VIL : Left tip of linkage group VI
VIL. Left of chol-2.
Tel-VIL is linked to RFLP markers in VIL (1447, 1810).
Tip-VIR : Right tip of linkage group VI
VIR. Marked by quasiterminal translocations T(UK-T12), T(NM103), and T(ALS159), which are right of ws-1, col-18, un-23, and trp-2 (13%) (1578, 1582).
Tel-VIR is linked to RFLP markers in VIR (64, 1447, 1809).
Tip-VIIL : Left tip of linkage group VII
VIIL. Left of cya-8 and T(ALS179).
Tel-VIIL is linked to RFLP markers in VIIL (1447, 1810).
Tip-VIIR : Right tip of linkage group VII
VIIR. Right of sk.
tk : thymidine kinase
Introduced from herpes simplex virus.
Fungi are deficient in the synthesis of thymidine kinase. The absence of endogenous thymidine kinase activity in wild-type Neurospora makes tk a readily detectable reporter gene. Expression of the viral thymidine kinase gene makes it possible to label DNA efficiently with radioactive thymidine and with fluorodeoxyuridine (1772). Strains expressing TK become sensitive to nucleoside drugs that are activated by TK (1772). Strains heterokaryotic for tk can therefore be resolved by using fluorodeoxyuridine to select conidia that contain no tk-bearing nuclei (1338).
Tn5 : Transposon 5
Introduced from Escherichia coli.
Used for insertional mutagenesis (1269); see also ref. (823).
tng : tangerine
IIL. Between pyr-4 (16%), T(NM149) and arg-5 (6%; 14%). Probably left of thr-2 (2%) (1582).
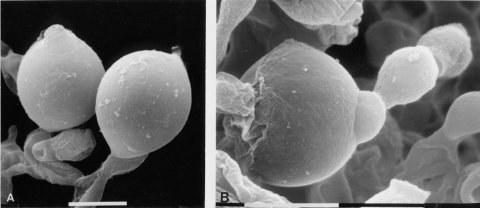
Irregular spreading growth. Hyphae are not curled (unlike ro) and are not as densely branched as those of col-4. Female infertile. Giant conidia are formed (1582) (Fig. 63), which may have many nuclei (1673). Some proconidia continue to grow beyond the normal 5- to 10-mm diameter of wild type to 100 mm or more, engulfing neighboring conidia and hyphae (1958). Microcapillary electrodes can be inserted in the spherical giant conidia and used to determine membrane potentials and resistances (187).

FIGURE 63 Giant conidia of the mutant tng: tangerine. A large conidium in the photograph to the right has partially engulfed a normal size conidium in the same conidial chain. Bar length = 10 mm. Scanning EM photographs from M. L. Springer. The photograph on the left is from ref. (1958 ), used here with permission from the Cold Spring Harbor Laboratory Press.
tnr : tetrahydroxynaphthalene reductase
VIIL. Between nic-3 (3T/18 asci) and ars-1 (2T/18 asci), Cen-VII (3T/18 asci) (1452).
Cloned and partially sequenced: GenBank AA902051, AA902055 EST NM6H10 (549).
Encodes putative tetrahydroxynaphthalene (T4HN) reductase (EC 1.1.1.252) (549).
tns : tenuous
VIIR. Between T(5936) and arg-10 (1589).
Growth is thin and severely retarded. Recovered as a putative RIP allele in progeny from Dp(VIIR®IL)5936 ´ Normal (1589).
tol : tolerant
IVR. Between his-5 and trp-4 (1%) (1466, 1911).
Cloned and sequenced: EMBL/GenBank AF085183; Orbach-Sachs clone X25D7.
A key regulatory gene that controls mating-type-mediated vegetative incompatibility. The tol gene does not affect sexual compatibility. mat A + mat a heterokaryons are vegetatively incompatible in standard wild-type Neurospora crassa. If the normal N. crassa allele, designated here tolC, is replaced by a recessive allele, tol, heterokaryons of constitution tol mat A + tol mat a are fully compatible and stable, provided that other het loci are homokaryotic. Growth of mat A/mat a partial diploids is inhibited and highly abnormal when the dominant normal allele tolC is present, but mat A/mat a duplications grow normally when tolC is replaced by a recessive tol allele (1466). Vegetative incompatibility mediated by het genes other than mating type is not affected by the presence or absence of tolC (1175, 1466, 1560). Mutation or deletion of tolC restores normal growth rate and morphology to slow-growing, unstable, mixed-mating-type (tolC mat a + tol mat A) heterokaryons (517). A recessive tol allele is present in some isolates from nature, and recessive alleles have arisen spontaneously at least twice in laboratory stocks (1466, 1582, 2277). Seven mutants selected as spontaneous escapes from inhibited growth of mat A/mat a partial diploids were all tol alleles (2146). Double mutant tol trp-4 stocks are convenient because the closely linked trp-4 tags the tol allele, which otherwise requires progeny tests for scoring. tol has been used as an alternative to mat am33 for maintaining stable mat A + mat a heterokaryons and allowing a disadvantaged component to be used as parent in a cross (1456). Homozygous tol may partially restore fertility to crosses involving fmf-1, which otherwise produce no fertile perithecia (1003). Mutations that inactivate tolC were interpreted as recessive suppressors of the mating-type-mediated vegetative incompatibility. In Neurospora tetrasperma, which normally grows as a vegetatively compatible matA + mat a heterokaryon, substitution of the N. crassa tol allele (tolC) for the N. tetrasperma allele (tolT) makes the N. tetrasperma mating types vegetatively incompatible, whereas the reverse substitution of tolT for tolC in N. crassa makes the N. crassa mating types vegetatively compatible (977). The normal allele in N. tetrasperma, tolT, resembles the N. crassa recessive mutant tol. In both species, the vegetative incompatibility mediated by mat A and mat a thus depends on tol function, and suppression is due to the absence of tol function.
tom : translocase of outer mitochondrial membrane
Translocase of the outer mitochondrial membrane (1612). Reviewed in ref. (1459). Involved in the translocation of nuclear-coded preproteins to the mitochondrial compartment. Subunits of the complex include TOM70, TOM40, TOM22, TOM20, TOM7, and TOM6 (Fig. 64). The 13.8-nm TOM complex particle has been purified and its structure and subunit stoichiometry analyzed (1112). Subunits were formerly called MOM (1612).
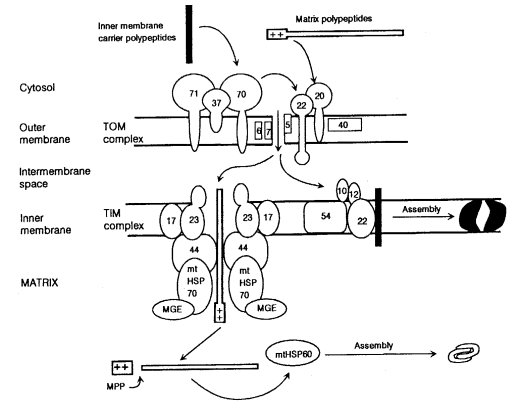
FIGURE 64 Import pathways of mitochondrial polypeptides synthesized in the cytosol. The TOM complex consists of surface-exposed components that function as preprotein receptors and of membrane-embedded components that are involved in passing the preproteins across the mitochondrial outer membrane. The TIM complexes deal with polypeptides having different final destinations. The peripheral protein TIM44 provides a docking site for HSP70 at the inner membrane. The products of the nuclear genes hsp60 and hsp70 are called mtHSP60 and mtHSP70 in the figure. MPP: mitochondrial processing peptidase. From ref. (461 ), [Davis, R.H. (2000). "Neurospora: Contributions of a Model Organism."], where it was redrawn from a figure provided by Frank Nargang. Reproduced with permission from Oxford University Press.
tom6 : translocase of outer mitochondrial membrane 6
Unmapped.
Cloned (1112).
Encodes the 6-kDa component of the translocase. Considered to be part of the translocation pore (1112, 1459); interacts with TOM40 (1700). Formerly known as mom8 (29, 1945) (Fig. 64).
tom7 : translocase of outer mitochondrial membrane 7
Unmapped.
Cloned (1112).
Encodes the 7-kDa component of the translocase. Considered to be part of the translocation pore (1112, 1459). Formerly known as mom7 (1049, 1373, 1945) (Fig. 64).
tom20 : translocase of outer mitochondrial membrane 20
IVR. Between pyr-1 (2T/18 asci) and trp-4 (4T/18 asci). Linked to pho-5 (0T/18 asci) (1447).
Cloned and sequenced: Swissprot OM20_NEUCR, EMBL/GenBank M80528, EMBL NCMITOM, GenBank NEUMITOM.
Specifies product originally identified as a 19-kDa outer mitochondrial component (Fig. 64). Antibodies directed against TOM20 block the import of many mitochondrially destined, nuclear-encoded proteins (1943), but it is not essential for ADP/ATP carrier protein or cytochrome c import (840). Required for TOM22 import (1039). Sheltered RIP showed that tom20 inactivation results in very slow growth and multiple mitochondrial defects (839, 840). Homokaryons lacking tom20 have extremely slow stop-start growth (839). Formerly known as mom19.
tom22 : translocase of outer mitochondrial membrane 22
VIL. Linked to gdh-1 (0T/16 asci), nuo19.3 (9T/18 asci) (1447).
Cloned and sequenced: Swissprot OM22_NEUCR, EMBL/GenBank X71021, GenBank NCMOM22, PIR A40669.
Specifies an essential 22-kDa component of the mitochondrial protein import complex (1439). TOM22 is anchored to the outer membrane through a single membrane-spanning domain (1049) (Fig. 64). Functions with TOM20 as the preprotein receptor (415, 1049, 1303, 1701). A positively charged internal region of TOM22 just prior to the membrane-spanning domain has a role in targeting the TOM22 polypeptide to the outer mitochondrial membrane (1735) . Abundant negative charges in the cytosolic domain may not be important for making TOM22 competent for function in translocation (1440). TOM22 influences TOM40 oligomerization (1700). It is required for the import of most mitochondrially destined proteins, including the ADP/ATP carrier. Called mom22.
tom40 : translocase of outer mitochondrial membrane 40
VR. Linked to cyh-2 (0T/15 asci) (1447).
Cloned and sequenced: Swissprot OM40_NEUCR, EMBL/GenBank X56883, GenBank NCMOM38, PIR S13418.
Encodes TOM40, a major component of the translocation pore (1040, 1050), which also consists of TOM6 and TOM7 (1049, 1945) (Fig. 64). TOM40 is closely associated with polypeptides as they are translocated across the mitochondrial outer membrane (1701, 1945). Sheltered RIP shows tom40 to be essential (1436), as is tom22 (1439). Called mom38.
tom70 : translocase of outer mitochondrial membrane 70
VR. Right of inl (6 or 7 T/17 asci) (1447).
Cloned and sequenced: Swissprot OM70_NEUCR, EMBL/GenBank X53735, AF110494, PIR A36682, GenBank NCMOM72.
Identified as encoding an outer mitochondrial membrane protein required for the import of the ATP/ADP carrier (1944, 1998) (Fig. 64). Only a fraction of total mitochondrial TOM70 associates with the TOM complex (1112). Mutants obtained by RIP are viable with a slow-growth phenotype exacerbated at elevated temperatures, and they conidiate poorly; mitochondria lacking TOM70 import the ADP/ATP carrier poorly (761). Called mom72.
Tp : Transposition
The name Transposition is used for intrachromosomal rearrangements in which an interstitial segment that contains two or more contiguous genes is removed from its original position and inserted elsewhere in the same chromosome. In crosses of Transposition ´ Normal, crossing over may produce duplications and deficiencies for defined chromosome segments and may result in dicentric bridges and acentric fragments, depending on the orientation and position of the transposed segment relative to the centromere (1605). The symbol for transpositions resembles that for insertional translocations, but with Tp in place of T.
tru : transport of uracil
Allelic with uc-5.
Transport
Amino acid transport across the plasma membrane is controlled by genes in three systems: basic (pmb, =bat); neutral (mtr, =pmn), and general (pmg; see also su(mtr)-1) (Fig. 47). Genes argR, lysR, hlp-1, and hlp-2 were described as being involved in the transport of amino acids. Mutants that affect (or may affect) the transport of other metabolites or ions include car, cys-13, cys-14, fpr, glt, ipm-1, ipm-2, mea-1, sit, sor, trk, tys, uc-5, and ud-1. Mutants that appear to affect more than one transport system include cpc, fpr-6, hgu-4, nap, and un-3; see ref. (2230). General regulatory loci such as nit-2, cys-3, nuc-1, nuc-2, preg, and pgov control classes of related enzymes, including the relevant permeases (Figs. 24, 49, 50, and 51). Other genes concerned with uptake are acu-16 (acetate), pho-4, pho-5 (phosphate), and puu (putrescine). Older literature on the transport of various ions and compounds is reviewed in refs. (769) and (1799), amino acid transport in ref. (2231), and peptide transport in ref. (2232). Nuclear genes called tom and tim specify subunits of translocases associated with the outer and inner membranes of mitochondria, respectively (Fig. 64). The por gene mediates the transport of metabolites into the mitochondrion. Ornithine transport across the mitochondrial membranes is mediated by arg-13 (Fig. 11). See pma for the transport of protons across the plasma membrane and vma for the transport of protons across the membranes of mitochondria and other intracellular components (Fig. 66).
Transposable elements
The retrotransposon Tad, which was discovered in a single wild strain, is the only known active transposable element in Neurospora. Inactive relics of numerous transposable elements are present in the genome, however (see Dab-1,Guest, Pogo, Punt, Tad, Tcen, Tg11, Tg12). Centromere regions are rich in copies of defective transposable elements (see Cen-VII). For reviews of transposons in filamentous fungi, see refs. (439) and (1044). The paucity of active elements in Neurospora, in contrast to their demonstrated abundance in ameiotic filamentous fungi (439), is attributed to the RIP process, which inactivates duplicated sequences in Neurospora by inducing G:C to A:T mutations prior to meiosis. Consistent with this hypothesis, many of the inactive elements bear earmarks of having undergone RIP.
tre-1 : trehalase-1
IR. Between met-6 (7%) and ad-9 (10%) (2025, 2210, 2286).
Unable to use trehalose as the carbon source. Structural gene for acid trehalase (EC 3.2.1.28) (2210). The Neurospora enzyme is classified as an acid trehalase of a subclass that is insensitive to Ca2+ and Mn2+ and to inhibition by ATP [reviewed in ref. (1004)]. Mutations affect thermostability and the production of an inhibitor of the wild-type enzyme (2025, 2210). Also, the invertase level is reduced by 50% and amylase is increased. The trehalase level is reduced to 10% of the wild type in (tre + tre+) heterokaryons and to 1% in tre/tre+ duplications. Used in studying trehalose metabolism (190, 524). For a possibly related regulatory mutation, see ref. (1323). Laboratory stocks of Neurospora crassa and wild isolates of Neurospora intermedia are polymorphic for trehalase electrophoretic mobility; one such variant was called mig (2025, 2210, 2286).
tre-2 : trehalase-2
Unmapped.
Cloned and sequenced: EMBL/GenBank AF044218.
Encodes neutral trehalase (α,α-trehalose glucohydrolase) (EC 3.2.1.28) (524). Obtained by PCR.
trk : transport of potassium
IIIR. Linked to leu-1 (0/92) (1926).
Cloned and sequenced: GenBank AJ009758.
Encodes a major potassium transporter, homologous to the TRK transporters of Saccharomyces cerevisiae (842). Requires high K+ concentration for growth. Na+ cannot substitute. Cation transport system maximum velocity is normal; Km is 3 times normal. Recessive. Obtained by inositolless death enrichment on low potassium (1926).
tRNALEU : leucine tRNA
VIIL. Between ars-1 and the qa cluster, adjoining qa-1F (938).
Cloned and sequenced: EMBL/GenBank X00736, GenBank NCTRNLEU. Included in VII contig GenBank AC006498.
Encodes leucine tRNA, with the AAG anticodon. The gene contains an intervening sequence (938).
tRNAPHE : phenylalanine tRNA
Unmapped.
Cloned and sequenced: EMBL/GenBank X02710, J01251, GenBank NCTRNPHE, NEUTGFF3.
Encodes phenylalanine tRNA. Contains a 16-bp intron adjacent to the anticodon (1884).
tRNA Synthetase
Cloned genes that specify tRNA synthetases include cyt-18, cyt-20, leu-5, leu-6, and un-3. The activity of Trp tRNA synthetase is greatly reduced in the trp-5 mutant.
trp : tryptophan
For the aromatic biosynthetic pathway, see Fig. 13. Tryptophan is required in much higher concentrations than its precursors, anthranilic acid and indole. A concentration of 0.01 mg/ml precursors is sufficient, but up to 0.1 or 0.2 mg/ml tryptophan is required by some mutants. High concentrations of anthranilic acid are toxic. Most trp mutants grow better on 0.2 mg/ml tryptophan plus 0.2 mg/ml phenylalanine than on tryptophan alone. Also called tryp, try.
Regulation: Tryptophan feedback inhibits anthranilate synthase, anthranilate-PP-ribose P-phosphoribosyl transferase, and one of three isozymes of DAHP synthase (the first step in aromatic biosynthesis). Tryptophan stimulates chorismate mutase, directing chorismate to prephenate rather than to anthranilate synthesis. All four genes of the tryptophan pathway are derepressed by starvation for tryptophan. High indoleglycerol phosphate levels also cause derepression. Derepression may involve inhibition of trp tRNA synthetase (87, 318, 483, 821, 1442, 1961). Genes for tryptophan biosynthesis are derepressed coordinately with those for histidine, arginine, and lysine biosynthesis; this is called "cross-pathway regulation" (292). Reviewed in refs. (1281) and (1768); see cpc.
trp-1 : tryptophan-1
IIIR Between ad-2 (1%; 7%) and ro-2 (2%; 12%) (18, 426, 1585). Linked to fpr-3 (<1%) (1057).
Cloned and sequenced: Swissprot TRPG_NEUCR, EMBL/GenBank J01252, PIR A01130, NNNC2, EMBL NCP1, GenBank NEUTRP1.
Structural gene for three domains and activities: anthranilate synthetase component II (EC 4.1.3.27) (the glutamine amino transferase activity; trp-2 specifies component I of this activity), phosphoribosylanthranilate (PRA) isomerase (EC 5.3.1.24), and indoleglycerolphosphate (InGP) synthetase (EC 4.1.1.48) (347, 484) (Fig. 13). Reviewed as an example of gene fusion (425). Uses tryptophan or indole (2061). Some alleles can also use anthranilate, but others cannot (11). Alleles differ in lacking one or more of the three activities. For example, trp-1 allele (15) lacks all three activities; trp-1 allele (20) lacks only PRA isomerase, trp-1C (1) lacks only anthranilate synthetase, trp-1 allele (25) lacks both PRA isomerase and InGP synthetase, etc. (523). To avoid confusion, note that in ref. (523) and related papers, the same "allele number" may be used for a trp-2 mutant, a trp-1 mutant (anthranilate-non-utilizing) and a trp-1C mutant (anthranilate-utilizing); mutants of the last class are listed by FGSC as trp-1 with the allele number prefixed by C. Alleles differ in their ability to form aggregates (347, 523). The Neurospora trp-1 gene is only partially expressed in Escherichia coli. Fine-structure maps (17, 523). Complementation maps (17, 330). A nonsense (amber) allele was used to demonstrate the restoration of normal enzyme aggregate by supersuppressors (348). Alleles that accumulate anthranilate are scorable by blue fluorescence under long-wave UV after 2-5 days growth on minimal medium + indole (10 mg/ml) at 34ºC (1587, 1592). Aging cultures may produce brown pigment; the blue fluorescence disappears as pigment forms.
trp-2 : tryptophan-2
VIR. Between del (0; 13%), T(OY329) and un-23 (5%; 27%), T(OY320), ws-1 (38%) (1578, 1603, 1604, 1992).
Cloned (257): pSV50 clone 2:5D.
Structural gene for anthranilate synthase component I (EC 4.1.3.27) (1038) (Fig. 13). Uses kynurenine, anthranilic acid, indole, or tryptophan (191). Kynurenine is utilized by conversion to anthranilate (846). TRP-2 catalyzes anthranilate synthesis with ammonia, but not with glutamine as the amino donor (62). Anthranilate synthetase (glutamine-linked) is specified in collaboration with trp-1 in the trifunctional TRP-1+ TRP-2+ enzyme aggregate (347, 523) (Fig. 13; see trp-1 entry). A nonsense (amber) allele was used to isolate supersuppressors (1859) and to study the enzyme complex restored by supersuppressors (348).
trp-3 : tryptophan-3
IIR. Between fl (2%; 6%), mus-23 (6%, 9%) and un-5 (10%), rip-1 (9%) (85, 1582, 1592, 1828, 2190).
Cloned and sequenced: Swissprot TRP_NEUCR, EMBL/GenBank J04594, PIR A28162, A32959, EMBL NCTRP3A, GenBank NEUTRP3A; pSV50 clone 5:4G.
Structural gene for tryptophan synthetase (EC 4.2.1.20) (2264) (called tryptophan desmolase in early literature). Uses tryptophan (1362); some alleles also use indole (11). Tryptophan synthetase catalyzes three reactions: indoleglycerol phosphate (InGP) ® tryptophan, indole ® tryptophan, and InGP ® indole (Fig. 13). In Neurospora, all three reactions are catalyzed by a single trifunctional protein, which is specified by a single gene (1289, 2264). Mutants lack InGP ® tryptophan activity, but differ with respect to the other activities. For example, trp-3td141 is blocked in InGP utilization but can use indole; trp-3td100 can synthesize indole but cannot convert it to tryptophan, and trp-3td140 is a polar mutant lacking all three activities. [See refs. (2029) and (1127) for references and characteristics of other mutants.] Used extensively for studies of gene structure in relation to enzyme activity (482, 1127, 2264). The active enzyme is a homo-oligomer (1289); the monomer has two domains (257, 1288, 1290). Biochemical studies of complementation between alleles in vivo (1127, 1128) and in vitro (2028). Complementation maps (11, 12, 16, 1127). Fine-structure maps (12, 1024, 1127, 2029). Reviewed as an example of gene fusion (425). trp-3 mutant C83 provided the first proved example in Neurospora of gene-controlled loss of enzyme activity (1362); trp-3 mutant S1952 provided the first example of allele-specific suppression restoring a functional wild-type-like enzyme (2262). Allele td140 is an amber mutant suppressible by ssu-1 (1859, 1860). Certain classes of trp-3 mutants are osmotic-remediable (1128). Called td, tryp-3.
trp-4 : tryptophan-4
IVR. Between his-5 (3%; 7%), tol (1%) and leu-2 (1% or 2%) (103, 1255, 1466, 1911, 1928).
Cloned: pSV50 clone 32:2G, Orbach-Sachs clones X4E10, X10A04, X17C11, G6C09, G6E12.
Uses tryptophan or indole (1477). Deficient in anthranilate-PP-ribose-P-phosphoribosyl transferase (EC 2.4.2.18) (2199) (Fig. 13). Scorable by blue fluorescence (anthranilate) in medium under long-wave UV after 2-5 days growth on minimal medium + indole (10 mg/ml), at 34oC. Initial stocks of the first trp-4 mutant were inhibited by suboptimal concentrations of tryptophan (1477), but derivatives have been obtained that are free of this problem (1973).
trp-5 : tryptophan-5
VR. Between inl (4%), pab-1 and met-3 (4%) (13, 21, 1582).
Uses anthranilate, indole, or tryptophan (21). Tryptophanyl-tRNA synthetase activity (EC 6.1.1.2) is 5% that of wild type (1442). Anthranilate synthetase and tryptophan synthetase are derepressed (1442). Not temperature-sensitive.
trx : thioredoxin
III. Present in the same DNA fragment with acr-2 but transcribed in the opposite direction (27).
Cloned and sequenced: Swissprot THIO_NEUCR, EMBL/GenBank D45892, EMBL NCTRX, GenBank NEUTRX.
Encodes a protein having extensive homology to thioredoxins from various organisms (27).
try : tryptophan
Changed to trp.
tryp : tryptophan
Changed to trp.
ts : tan spore
VR. Linked to inl (4%) (1428).
Ascospores are slow to mature, remaining light brown when wild-type ascospores have blackened. Expressed autonomously, allowing visual scoring in heterozygous asci. Only a small minority of ts ascospores, those that have darkened with age, are capable of germination. Photograph of asci (1428). Used to study multiple (1431) and selective (597) fertilization, preferential segregation (1429), and factors affecting crossing-over frequency (1138, 1430).
tu : tuft
IIR. Between pe (8%) and fl (19%) (1203).
Conidia are formed mostly in clusters at the shallow end of agar slants (1203). One of the first Neurospora genes to be mapped (stock lost).
tubR : tubercidin resistant
Unmapped. Unlinked to pyr-1 (1244).
Selected for resistance to tubercidin (7-deaza-adenosine), a competitive inhibitor of adenosine (1244). Adenosine kinase activity is reduced to ~10% that of wild type. Transport of adenosine into the cell is normal. No mutant stock survives (1244).
Tubulin
See Bml, tba, and tbg.
tub-2 : tubulin-2
Has sometimes been used for Bml, which has precedence.
ty : tyrosinase
The symbol ty is used for genes that regulate tyrosinase. The structural gene for tyrosinase is symbolized T. Mutants at all of these loci are prototrophic, growing on unsupplemented minimal medium.
ty-1 : tyrosinase-1
IIIR. Linked to tyr-1 (0%; 6%), dow (21%) (1582, 2179).
Tyrosinase is repressed. Recessive (899, 901). Uninducible by inhibitors of protein synthesis in the sexual phase, but inducible in vegetative culture (898, 901). Prototrophic. Velvetlike vegetative morphology. Infertile as female (no or few perithecia), but fertile as male. Infertility is recessive in a heterokaryon (899). Low ascospore viability (880). Scored by color reaction with DL-DOPA as substrate (899, 901) or by morphology and female sterility.
ty-2 : tyrosinase-2
IR. Right of al-2 (880).
Tyrosinase is repressed. Prototrophic, recessive (899, 901). Inducible in vegetative culture by protein-synthesis inhibitors such as cycloheximide, but uninducible in the sexual phase (898, 901). Described as having short aerial hyphae (880), although earlier said to be morphologically normal (899). Female infertile, but fertile as male. Infertility is recessive in a heterokaryon (899). Scored by color reaction with DL-DOPA as substrate (899, 901), or by morphology, and female sterility.
ty-3 : tyrosinase-3
IIIR. Between Cen-III and ad-4 [W. L. Chan, Ph. D. Thesis, University of Malaya, Kuala Lumpur, 1977, cited in ref. (881)]. Not allelic with T or ff-3 [368; N. H. Horowitz and H. MacLeod, cited in ref. (623)].
Tyrosinase is repressed. Uninducible in vegetative culture by inhibitors of protein synthesis such as cycloheximide (623, 881). Originally reported to be female-sterile and morphologically abnormal. These properties were later shown to be due to a second, nonallelic mutation, ff-3. The ty-3 single mutant is female-fertile and morphologically normal (368). Scored by color reaction with DL-DOPA as substrate (899, 901). The original strain, called T22, contained ty-4 in addition to ty-3 and ff-3.
ty-4 : tyrosinase-4
Unmapped.
Tyrosinase repressed. Uninducible by total starvation in sodium phosphate buffer, but can be induced by cycloheximide. Found in strain T22, which also contains ty-3 and ff-3 [W. L. Chan, Ph. D. Thesis, University of Malaya, Kuala Lumpur, 1977, cited in ref. (881)]; also present in some Emerson wild types (881).
tyr-1 : tyrosine-l
IIIR. Between vel (3%; 5%), phe-2 (2%; 4%) and un-17 (4%) (601, 1322, 1582, 1592).
Requires tyrosine (2057). Lacks prephenate dehydrogenase activity (EC 1.3.1.13) (87, 601) (Fig. 13). Shows phenotypic adaptation after a lag, attaining wild-type growth rate on minimal medium. Adaptation is not carried through conidia (2057). Inhibition of prephenate dehydrogenase by phenylalanine examined (312). Allele UT145 was called tyr-3 (601).
tyr-2 : tyrosine-2
IR. Between T(4540)L nic-2 and T(Y112M4i)R, ace-7, cr-1 (2%) (1578, 1585).
Requires tyrosine (1585). Prephenate dehydrogenase activity is decreased (601). pe-like morphology. Female sterile. tyr- ascospores do not become fully pigmented. Growth is suboptimal even on fully supplemented medium. Adapts to growth on minimal medium after several days. Scoring by growth on slants is treacherous, but the mutant can be recognized by its darkening of tyrosine-supplemented minimal medium.
tyr-3 : tyrosine-3
Used for an allele of tyr-1.
tyr-s : tyrosine sensitive
Symbol changed to tys.
tys : tyrosine sensitive
I. Right of mat (6%) (1582, 2234).
Growth is inhibited 80% by 0.07 mM L-tyrosine. Growth on minimal medium also is inhibited by glycyl-leucyl-tyrosine and by various tyrosine analogues. Uptake of L-[14C]-p-tyrosine is increased slightly, but this is not proposed as the cause of the inhibition. The primary defect is unknown (2234). Used to obtain the oligopeptide transport mutant glt, which is resistant to glycyl-leucyl-tyrosine but not to tyrosine (2236). Called tyr-s (2234, 2236).
Return to the 2000 Neurospora compendium main page