vac-5 : vacuolar protein-5
Changed to htl.
val : valine
VR. Right of at (10%). Linked to ilv-2 (0/135) (1582).
Alleles 33026 and 33050 appear to be valine auxotrophs, not requiring or responding to added isoleucine (907, 1582). Not defective in valyl-tRNA synthetase (610), as are the only val mutants known in bacteria; it may therefore define a gene specifying the unique conversion step from α-keto-isovalerate to valine, branched-chain amino acid aminotransferase (EC 2.6.1.42). The requirement is somewhat leaky, not temperature-sensitive. An ilv mutant strain with an incomplete isolation number, ilv(?6201), was incorrectly designated val (45201), and was the source of linkage data for a locus erroneously shown as val on maps prior to 1980.
van : vanadate resistant
Changed to pho-4.
var-1 : variant-1
Unmapped. Segregates as a single gene (169).
The rate of growth is reduced by 40% and aerial hyphae are decreased, resulting in a shaved appearance. Functional protoperithecia are absent. Lysed areas appear and spread in old cultures. Not rejuvenated in heterokaryons. Originated in an experiment on uninterrupted growth (169).
vel : velvet
IIIR. Between ro-2 (6%), T(D305)L and tyr-1 (3%; 5%). Linked to phe-2 (l%) (1582, 1587, 1592).
Soft, colonial growth (1548). Flat at first and then elevated, with conidia sometimes formed later, in aerial puffs. Good female fertility. col-13 (R2471) and col-15 (R2531) are putative alleles. Mutations in strains R2471 and R2531, named col-13 and col-15 in ref. (719), are probably vel alleles, resembling vel in phenotype and mapping in the same IIIR region [see entries in ref. (1596)].
Inseparable from translocation T(I;III)3717 (1578). Aconidial flat morphology (1580). No corresponding point mutant is known.
vma : vacuolar membrane ATPase
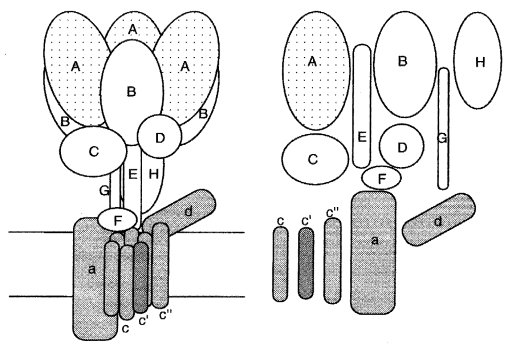
This multiple-subunit complex acts as a proton pump to acidify the vacuole and related intracellular compartments (208). The vacuolar membrane ATPase complex is organized into a hydrophobic sector, V0, that conducts protons through the membrane, and a protruding hydrophilic sector, V1, that hydrolyzes ATP. Reviewed in ref. (1271) (Fig. 66).

FIGURE 66 A diagram of the vacuolar membrane ATPase. Subunits in the hydrophilic portion have been given capital letters and those associated with the membrane lower-case letters. Symbols: A, Vma-1; B, Vma-2; D, Vma-8; E, Vma-4; F, Vma-7; G, Vma-10; H, Vma-13; a, Vph-1; c, Vma-3; c , Vma-11; d, Vma-6 (1271 ).
vma-1 : vacuolar membrane ATPase-1
VR. Between sp (8%) and am (2%; ~40 kb). Linked to vma-3 (~100 kb), cyh-2, leu-5 (0T/17) (1447, 1920).
Cloned and sequenced: Swissprot VATA_NEUCR, EMBL/GenBank J03955, EMBL NCVMA1A, GenBank NEUVMA1A; pSV50 clone 14:4A.
Specifies vacuolar ATPase (EC 3.6.1.3), 67-kDa subunit (subunit A) (215) (Fig. 66). This subunit contains the site at which ATP is bound and hydrolyzed. Although the vma-1+ gene initially appeared to be essential (628), mutants disrupted by RIP and lacking detectable vacuolar ATPase activity proved to be viable even though tip growth was inhibited, multiple branching was induced, and they were unable to differentiate conidia or perithecia (211a). The mutants were completely resistant to concanamycin C.
vma-2 : vacuolar membrane ATPase-2
IIR. Between preg (6T/18 asci) and Fsr-3 (1T/18 asci), eas (2T/18 asci) (1447).
Cloned and sequenced: Swissprot VATB_NEUCR, EMBL/GenBank J03956, EMBL NCVMA2A, GenBank NEUVMA2A.
Encodes vacuolar membrane ATPase (EC 3.6.1.3), 57-kDa subunit (subunit B) (Fig. 66). This subunit appears to contain an ATP-binding site, but nucleotides bound at this site are not hydrolyzed during catalysis (206).
vma-3 : vacuolar membrane ATPase-3
VR. Between his-1 and al-3. Linked to vma-1 (~100 kb), cyh-2, leu-5 (0T/17 asci) (1447, 1920).
Cloned and sequenced: Swissprot VATL_NEUCR, EMBL/GenBank L07105, EMBL NCVMA3A, GenBank NEUVMA3A; pSV50 clone 3:4G.
Encodes vacuolar ATPase (EC 3.6.1.3), 16-kDa Vo c-proteolipid subunit. This highly hydrophobic subunit is embedded in the membrane and probably forms part of the proton-conducting pathway through the membrane (1920) (Fig. 66).
vma-4 : vacuolar membrane ATPase-4
IL. Linked to nit-2 (0 or 1/16), Fsr-12 (0T/18 asci). Left of mat (4T/18 asci) (1447).
Cloned and sequenced: EMBL/GenBank U17641, EMBL NC17641; pSV50 clone 14:5H.
Specifies vacuolar ATPase (EC 3.6.1.3), 26-kDa E-subunit. This hydrophilic subunit may form part of the stalk that connects the hydrophilic ATP-binding sector with the integral membrane sector (214) (Fig. 66).
vma-6 : vacuolar membrane ATPase-6
II. Near Cen-II. In the same cDNA fragment with rap-1 (~1200 bp) (1317).
Cloned and sequenced: EMBL/GenBank U36470; Orbach-Sachs clone X2B08.
Encodes vacuolar ATP synthase (EC 3.6.1.3), 41-kDa subunit (Fig. 66). This polypeptide is tightly associated with the membrane, but does not appear to have any membrane-spanning hydrophobic regions (1317).
vma-7 : vacuolar membrane ATPase-7
IVR. Linked near Fsr-62 (2073).
Cloned and sequenced: GenBank AF099136; Orbach-Sachs clones G17C10, G20D10.
Encodes vacuolar ATPase (EC 3.6.1.34), 13-kDa F subunit (2073) (Fig. 66).
vma-8 : vacuolar membrane ATPase-8
IVR. Right of trp-4 (3T/18 asci). Linked to Fsr-13 (0T/18 asci) (1447).
Cloned and sequenced: EMBL/GenBank AF053230; Orbach-Sachs clone G15H03.
Encodes vacuolar ATP synthase (EC 3.6.1.34), 28-kDa D-subunit in the hydrophilic sector of the ATPase.
vma-10 : vacuolar membrane ATPase-10
IIR. Linked near arg-12.
Cloned and sequenced: EMBL/GenBank U84904; Orbach-Sachs clones G4E11, G6F12.
Encodes vacuolar ATPase (EC 3.6.1.3), 13-kDa G-subunit (Fig. 66). This subunit is in the hydrophilic sector and may function to connect the membrane-embedded and peripheral sectors of the enzyme (941).
vma-11 : vacuolar membrane ATPase-11
IR. Between al-2 (1T/18 asci) and arg-13 (5T, 2NPD/18 asci). Linked to sdv-4, -5 (0 or 1T/18 asci) (1447).
Cloned and sequenced: EMBL/GenBank AF162776; Orbach-Sachs clones X2A03, X2H07, G6B11, G25A04.
Vacuolar ATP synthase (EC 3.6.1.3), 16-kDa subunit. High sequence similarity to vma-3. This highly hydrophobic subunit is embedded in the membrane and probably forms part of the proton-conducting pathway through the membrane (1645) (Fig. 66).
vph-1 : vacuolar pH-sensitive ATPase-1
IIL. Left of Cen-II (5T/18 asci), Fsr-32 (4T/18 asci). Linked to Fsr-52 (0T/18 asci) (1447).
Cloned and sequenced: EMBL/GenBank U36396.
Encodes vacuolar membrane ATPase 98-kDa subunit. The N-terminal half of this polypeptide appears to be hydrophilic, and the C-terminal half has six or seven putative membrane-spanning regions (210) (Fig. 66). Named to conform with its yeast homologue, VPH1.
vvd : vivid
VIL. Between T(IBj5)L, ylo-1 and T(IBj5)R, cpc-1 (817, 1578, 1589). Not in a cosmid containing ylo-1 and lys-5 (1815).
Intense color. The level and speed of synthesis of carotenoids is elevated in al+ ylo+ strains, in the ylo-1 mutant, and in strains with pigmenting mutant alleles of the albino genes. Mutant vvd alleles have been obtained independently in three laboratories (573, 818-820, 1589). Expression of con-6 and con-10 in the light is increased when the vvd mutation is present (573).
Return to the 2000 Neurospora compendium main page