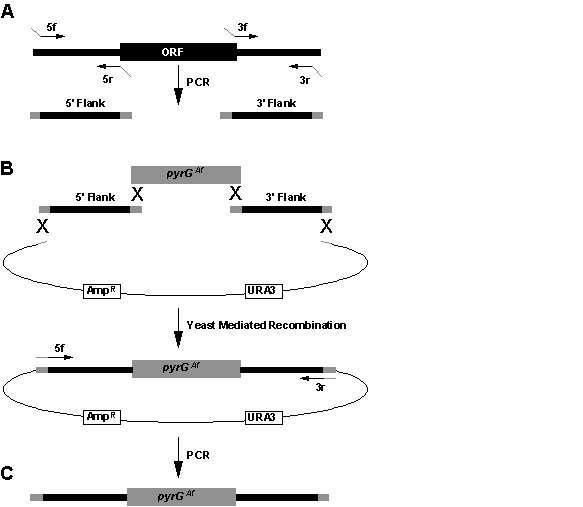

Figure
. Strategy for generation of deletion constructs. (A) Amplification of 5’ and 3’ gene specific flank fragments from A. nidulans genomic DNA. Primers 5r and 3f have 5’ extensions complementary to the pyrGAf cassette, whereas 5f and 3r have 5’ extensions complementary to the vector. (B) Cotransformation of yeast with the 5’ flank and 3’ flank together with the pyrGAf cassette and a gapped yeast shuttle vector generate a plasmid containing the gene specific deletion construct (Colot et al, 2006). (C) The final linear deletion construct is generated by PCR from the yeast DNA with primers 5f and 3r.Table Example primer set.
|
Primers for Anid_00038.1 |
|
|
5f |
GTAACGCCAGGGTTTTCCCAGTCACGACG ACTCCTGAGATGTCTCCTGT |
|
5r |
ATCCACTTAACGTTACTGAAATC GTAGCAGCGTGTTGACTACT |
|
3f |
CTCCTTCAATATCATCTTCTGTC GTTGGCAGGCCAGTAGTTT |
|
3r |
GCGGATAACAATTTCACACAGGAAACAGC TCTACCTGTACAGCATCAGC |
Listed are the gene specific primers for Anid_00038.1. The 5’ extensions are indicated in bold italics.
Reference
Colot HV
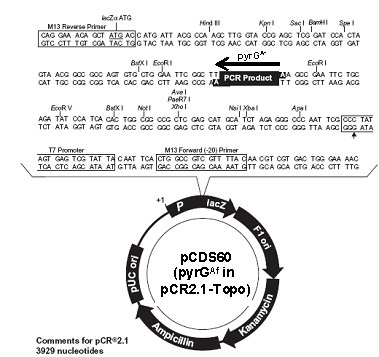
, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA 103:10352-10357.pCDS60
also available as an MS Word Document
10/08/10
The pyrGAf cassette was generated by PCR from pCDS60 using primers CDS164 and CDS165
pCDS60 insert:
EcoRI
CDS164 >GAATTC
GCCCTTGTTGGAGATTTCAGTAACGTTAAGTGGATCAAAGACATGATCTCTTACTGAGAGTTATTCTGTGTCTGACGAAATATGTTGTGTATATATATATATGTACGTTAAAAGTTCCGTGGAGTTACCAGTGATTGACCAATGTTTTATCTTCTACAGTTCTGCCTGTCTACCCCATTCTAGCTGTACCTGACTACAGAATAGTTTAATTGTGGTTGACCCCACAGTCGGAGGCGGAGGAATACAGCACCGATGTGGCCTGTCTCCATCCAGATTGGCACGCAATTTTTACACGCGGAAAAGATCGAGATAGAGTACGACTTTAAATTTAGTCCCCGGCGGCTTCTATTTTAGAATATTTGAGATTTGATTCTCAAGCAATTGATTTGGTTGGGTCACCCTCAATTGGATAATATACCTCATTGCTCGGCTACTTCAACTCATCAATCACCGTCATACCCCGCATATAACCCTCCATTCCCACGATGTCGTCCAAGTCGCAATTGACTTACGGTGCTCGAGCCAGCAAGCACCCCAATCCTCTGGCAAAGAGACTTTTTGAGATTGCCGAAGCAAAGAAGACAAACGTTACCGTCTCTGCTGATGTGACGACAACCCGAGAACTCCTGGACCTCGCTGACCGTACGGAAGCTGTTGGATCCAATACATATGCCGTCTAGCAATGGACTAATCAACTTTTGATGATACAGGTCTCGGTCCCTACATCGCCGTCATCAAGACACACATCGACATCCTCACCGATTTCAGCGTCGACACTATCAATGGCCTGAATGTGCTGGCTCAAAAGCACAACTTTTTGATCTTCGAGGACCGCAAATTCATCGACATCGGCAATACCGTCCAGAAGCAATACCACGGCGGTGCTCTGAGGATCTCCGAATGGGCCCACATTATCAACTGCAGCGTTCTCCCTGGCGAGGGCATCGTCGAGGCTCTGGCCCAGACCGCATCTGCGCAAGACTTCCCCTATGGTCCTGAGAGAGGACTGTTGGTCCTGGCAGAGATGACCTCCAAAGGATCGCTGGCTACGGGCGAGTATACCAAGGCATCGGTTGACTACGCTCGCAAATACAAGAACTTCGTTATGGGTTTCGTGTCGACGCGGGCCCTGACGGAAGTGCAGTCGGATGTGTCTTCAGCCTCGGAGGATGAAGATTTCGTGGTCTTCACGACGGGTGTGAACCTCTCTTCCAAAGGAGATAAGCTTGGACAGCAATACCAGACTCCTGCATCGGCTATTGGACGCGGTGCCGACTTTATCATCGCCGGTCGAGGCATCTACGCTGCTCCCGACCCGGTTGAAGCTGCACAGCGGTACCAGAAAGAAGGCTGGGAAGCTTATATGGCCAGAGTATGCGGCAAGTCATGATTTCCTCTTGGAGCAAAAGTGTAGTGCCAGTACGAGTGTTGTGGAGGAAGGCTGCATACATTGTGCCTGTCATTAAACGATGAGCTCGTCCGTATTGGCCCCTGTAATGCCATGTTTTCCGCCCCCAATCGTCAAGGTTTTCCCTTTGTTAGATTCCTACCAGTCATCTAGCAAGTGAGGTAAGCTTTGCCAGAAACGCCAAGGCTTTATCTATGTAGTCGATAAGCAAAGTGGACTGATAGCTTAATATGGAAGGTCCCTCAGGACAAGTCGACCTGTGCAGAAGAGATAACAGCTTGGCATCACGCATCAGTGCCTCCTCTCAGACAGGCTCCTTCAATATCATCTTCTGTCTCCGACAAGGGCGAATTC< CDS165 EcoRI
ATG = pyrGAf Start codon
TGA = pyrGAf Stop codon