An improved plasmid for transformation of Neurospora crassa using the pan-2 gene as a selectable marker
Wilson Low and Gregory Jedd.
Temasek Life Sciences Laboratory and Department of Biological Sciences,
National University of Singapore, Singapore 117604
gregory@tll.org.sg
Fungal Genetics Reports 55:44-45
(Pdf)
The pOKE103 plasmid can be used to transform Neurospora crassa using the pan-2 gene (NCU10048.3) as a selective marker. We mapped the pan-2 fragment to the right arm of LG VI, contig 4, and found that it carries a pan-2 linked gene, atg-12 (NCU10049.3). We report the removal of atg-12 to produce an improved plasmid, pOKE104.
Genetic transformation is a key tool to study fungi and the ability to create strains transformed with multiple plasmids is especially important for mechanistic cellular and developmental studies. In Neurospora crassa, a number of options are available: For reproducible expression, transgenes can be targeted as single copies to the his-3 locus by vectors designed to repair rarely reverting alleles at his-3 (Aramayo and Metzenberg, 1996; Margolin et al., 1997). Recently, a system has been reported, where integration at the csr-1 locus produces cyclosporin A resistance and allows the selection of single copy integrants at csr-1 (Bardiya and Shiu, 2007). Selection of transformants using Hygromycin B resistance is also common (Staben et al., 1989), however, the use of this marker for systematic deletion of Neurospora genes (Colot et al., 2006) is likely to preclude its use in many cases.
The pan-2 marker provides an additional means of transformation. In this case transformation is ectopic and results in the integration of single or multiple copies of the transgene. The pOKE103 vector constructed by J. Grotelueschen and Bob Metzenberg contains a genomic fragment of pan-2 in the pGEM3Z (+) vector backbone and this vector produces tight selection in pan-2 mutant backgrounds (Wu and Glass, 2001). We examined the pan-2 fragment and found that it carries an additional gene, atg-12, a conserved ubiquitin-related protein associated with cellular autophagy (Hanada and Ohsumi, 2005). Thus, transformation with pOKE103 may also result in variable overproduction of ATG-12. There is currently no evidence to suggest that transformation with pOKE103 produces any phenotypic transformation. Nevertheless, because the effects of additional copies of atg-12 may depend on the genotype and may emerge only in certain mutant backgrounds, we have removed atg-12 from pOKE103 to produce a simplified derived plasmid, pOKE104.
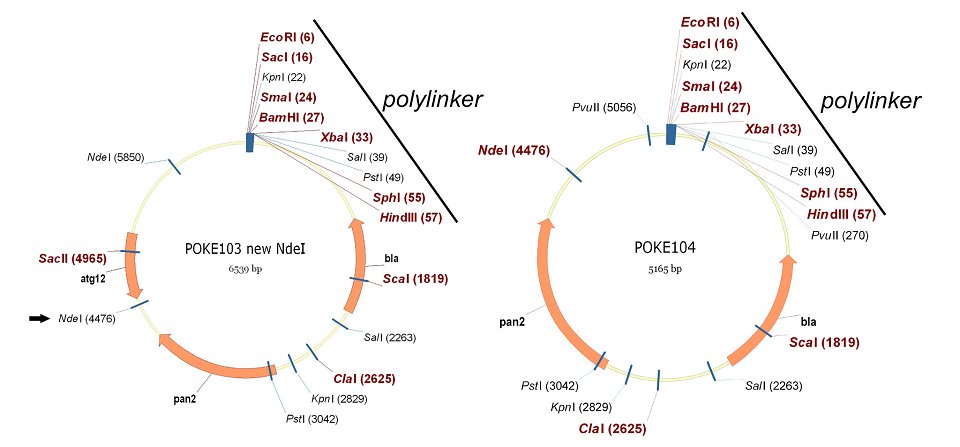
Plasmid pOKE 103 contains a unique NdeI restriction site 5' of the atg-12 gene and we used site-directed mutagenesis with two complementary primers ("new-NdeI"; GTC GGA ACA TAT GTT ATG GAC AGC ATG) to insert a second NdeI site 3' of atg-12 (Fig. 1). Digestion of this vector with NdeI removes a 1347 bp fragment containing the entire atg-12 gene. The vector fragment from this digestion was circularized to produce the pOKE104 vector (Fig. 1). The complete sequence of pOKE104 can be found using NCBI accession number FJ599664 and the plasmid can be obtained from the Fungal Genetics Stock Center (http://www.fgsc.net/).

Figure 1. Partial restriction maps (drawn with Vector NTI) of plasmids pOKE103 and pOKE104. The introduced NdeI site is shown on the left in pOKE103 (arrow); the right map shows pOKE 104 after removal of atg-12. The polylinkers are indicated with unique restriction sites shown in red bold.
Acknowledgements
We thank J. Grotelueschen and Bob Metzenberg for constructing the pOKE103 plasmid and the Temasek Life Sciences Laboratory for funding.
References
Aramayo, R., and R.L. Metzenberg. 1996. Gene replacements at the his-3
locus of Neurospora crassa. Fungal Genet Newslet. 43:9 -13.
Bardiya, N., and P.K. Shiu. 2007. Cyclosporin A-resistance based gene placement system for Neurospora crassa. Fungal Genet Biol. 44:307-14.
Colot, H.V., G. Park, G.E. Turner, C. Ringelberg, C.M. Crew, L. Litvinkova, R.L.
Weiss, K.A. Borkovich, and J.C. Dunlap. 2006. A high-throughput gene knockout
procedure for Neurospora reveals functions for multiple transcription factors.
Proc Natl Acad Sci U S A. 103:10352-7.
Hanada, T., and Y. Ohsumi. 2005. Structure-function relationship of Atg12, a
ubiquitin-like modifier essential for autophagy. Autophagy. 1:110-8.
Margolin, B.S., M. Freitag, and E.U. Selker. 1997. Improved plasmids for gene
targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal
Genet Newslet. 44:34-36.
Staben, C., B. Jensen, M. Singer, J. Pollock, M. Schechtman, J. Kinsey, and E.
Selker. 1989. Use of a bacterial Hygromycin B resistance gene as a dominant
selectable marker in Neurospora crassa transformation. Fungal Genet Newslet.
36:79-81.
Wu, J., and N.L. Glass. 2001. Identification of specificity determinants and
generation of alleles with novel specificity at the het-c heterokaryon
incompatibility locus of Neurospora crassa. Mol Cell Biol. 21:1045-57.