Results
DNA sequence analysis of 2.5 kb HindIII-NruI
fragment flanking the C- terminus of sid1
We sequenced the 2.5 kb HindIII-NruI fragment of U. maydis
strain 518 that is immediately downstream of sid1. Open reading frame
analysis revealed a single open reading frame encoding a protein of 547 amino
acids within the 1870 bp of sequenced DNA of the 2. 5 kb HindIII-NruI
fragment and corresponding to a BglII-NruI subregion. Gene
identity was determined by Blastn and tBlastx analysis using data available in
GenBank (www.ncbi.nlm.nih.gov). Analysis of
the translated sequence against translated sequences in Genbank revealed 50 hits
with significant sequence similarity (alignment score >= 200) for 56 accessions
representing genome sequences from fungi and bacteria. Multiple alignment of
the translated Ustilago gene with amino acid sequences of a putative Ade6
in the Ustilago strain 521 (Kamper et. al., 2006;
http://mips.gsf.de/genre/proj/ustilago/singleGeneReport.html?entry=um05162),
E. coli PurL and yeast Ade6 revealed significant similarity over
the regions compared. The N-terminal region of 547 amino acids of the
Ustilago strain 518 ade6 showed complete identity with the predicted
hypothetical protein for ade6 for Ustilago strain 521 in the MIPS
database. Sequence similarities of 54.3% with the translated purL gene
of E. coliB and 48.6% with the translated ADE6 gene of S.
cerevisiae were observed over this same region (data not shown).
Disruption of putative ade6 gene in U. maydis
Construction of plasmid pUC(2. 5HindIII-NruI):hygB. pUC(2.
5HindIII-NruI) DNA was digested at the single MluI site located near
the middle of the 2.5 kb HindIII-NruI fragment encoding the 5’
region of the putative Ustilago ade6 gene (Fig. 1). The
hygromycin resistance cassette was isolated as a HindIII fragment from
pHLI (Wang et al., 1989) after a double digestion with HindIII and
SspI. The single-stranded ends of the 3.4 kb HindIII fragment
encoding the hygromycin resistance gene were repaired with Klenow Polymerase and
cloned into the MluI site of pUC(2. 5HindIII-NruI) using T4
DNA ligase to produce the gene disruption construct.
Insert-containing clones were detected by hybridization of colony blots with a radioactively labeled hygromycin resistance gene probe. A restriction map of the cloned insert relative to the genomic region in which it is contained is shown in Figure 1.
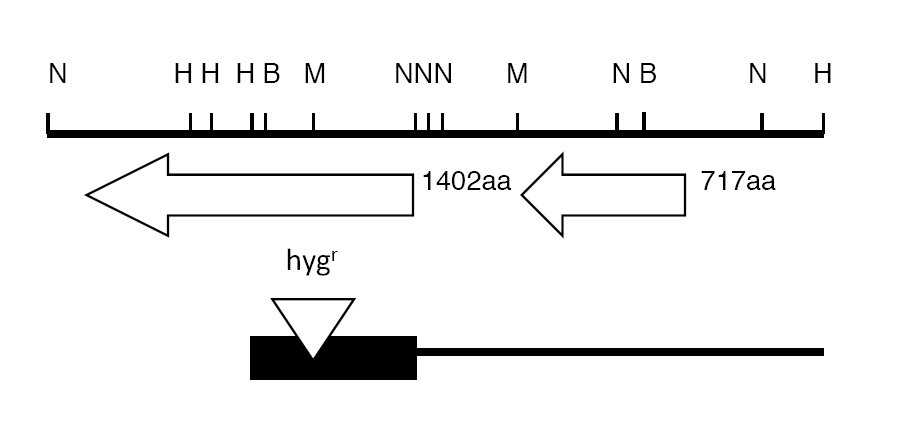
Figure 1. Partial restriction map of the ade6 –sid1 region of U. maydis. The HindIII-NruI fragment containing the hygromycin B resistance cassette inserted in the MluI site is shown at the bottom as a black box in relation to the 8.2 Kb HindIII fragment from which it was derived. Ade6 encodes a 1402 amino acid polypeptide while sid1 encodes a 717 amino acid polypeptide (Kamper et. al., 2007). Restriction sites shown are: B, BglII; H, HindIII; M, MluI; and N, NruI.
A HindIII/KpnI digest was carried out to linearize the ade6:hygB DNA for single-step gene disruption of U. maydis (Fig. 1). The KpnI site is part of the pUC18 polylinker and is one bp distal to the NruI:SmaI fusion site of the ade6:hygB construct. pHLI, used as a control, was linearized with HindIII. The plasmids were transformed into U. maydis 518. Transformants were selected on hygromycin-containing PDA medium.
|
Complete Medium |
Holliday, 1974 |
|
E-medium |
Wang et. al.,1989 |
|
Minimal Medium |
Holliday, 1974 |
|
E-medium + adenine (10 mg/L) |
Wang et. al., 1989 |
|
Minimal + adenine (10 mg/L) |
Wang et. al., 1989 |
Table 2. Media
Sixty hygromycin resistant transformants were transferred onto complete medium +
300μg/ml hygromycin (Table 2) and subsequently to minimal medium, minimal medium
+ adenine, E medium, and E medium + adenine. The resulting growth patterns of
eleven transformants were restricted on minimal and E medium, but were
unrestricted on minimal + adenine, E medium + adenine, and complete medium.
Southern hybridization analysis of the DNA of these 11 transformants using the
5’ half of the ade6 gene, and the hygromycin resistance gene, indicated
that homologous integration had resulted in disruption of the putative ade6
gene.
Segregation analysis was conducted on progeny obtained from three representative ade6 disruption mutants of Ustilago after crossing with wild type strain 521 containing the opposite mating type. The growth of progeny on minimal medium with and without adenine is summarized in Table 3. Data from crosses with Mutants 4 and 5 are not consistent with a single gene segregating for adenine auxotrophy, while that for Mutant 6 does suggest a single gene is present (p >>0.05). The possibility exists that some colonies chosen from the primary plating of germinated basidiospores were not derived from single spores, since the number of adenine prototrophs exceeds that of adenine auxotrophs. In mixed colonies on minimal medium, adenine prototrophs would mask the poor growth of adenine auxotrophs. Mutant progeny may also have grown more slowly or shown poorer viability on the primary plating medium thus skewing the data in favor of prototrophs.
|
Cross |
ade+ |
ade- |
X2 |
p |
|
Mutant 4 X 521 |
39 |
11 |
15.7 |
<0.00012 |
|
Mutant 5 X 521 |
33 |
17 |
5.1 |
0.0242 |
|
Mutant 6 X 521 |
27 |
23 |
0.32 |
0.57 |
Table 3. Segregation of adenine auxotrophy
in crosses of disruption mutants with wild type.1
1Degrees of freedom = 1
2Significant
Discussion
A hypothetical protein showing significant identity to
purL of E. coli and ADE6 of S. cerevisiae was
previously identified in the genome of U. maydis strain 521 (Kamper
et. al., 2006;
http://mips.gsf.de/genre/proj/ustilago/singleGeneReport.html?entry=um05162).
In this study the N-terminal region of the putative ade6 gene was
sequenced from strain 518 and disrupted by single step gene disruption. The
resulting transformants were incapable of growing on minimal and E media unless
supplied with adenine as would be expected for disruption of the U. maydis
formylglycineamide ribonucleotide synthetase gene. This gene marks one end of
the gene cluster encoding siderophore biosynthetic functions sid1 and
sid2. Our data thus provide functional confirmation of the designation of
the putative ade6 gene at position 308279 to 304071 of contig 1.188 on
Chromomsome 4 of the genome sequence of Ustilago maydis strain 521
(Kamper et. al., 2006;
http://mips.gsf.de/genre/proj/ustilago/singleGeneReport.html?entry=um05162).
Resistance to the drugs hygromycin and phleomycin has been widely used in Ustilago for selection of transformants (e.g., An et. al., 1997ab; Gold et. al., 1994). The auxotrophic strains generated in this study can be used as recipients for U. maydis transformation by complementation. In addition, ade6 mutants in combination with ade1 or ade2 mutants, that produce red pigments, can be used to design colony color screens for synthetic lethality and plasmid-generated mutations in a cloned gene (plasmid shuffle) (Sherman, 1998). The ade6 mutation prevents the formation of the red phosphoribosylamino-imidazole pigment; therefore, loss of the ade6-complementing plasmid leads to white sectoring in the colony. This method allows for direct visual analysis of desired genetic events without the need to replica plate. With the availability of self-replicating plasmids these methods should be readily adapted to Ustilago.
Acknowledgements
This work was supported by NIH grant GM33716 and USDA ARS CRIS project 3655-22000-013-00D to SAL. The authors also wish to thank Robin Holliday for strains 518 and 521 and Paige Taylor Breunig for her contribution to segregation analysis. DNA sequence of 1870 bp of the 2.5 kb HindIII-NruI fragment containing the 5’ portion of the U. maydis ade6 gene homolog was deposited in GenBank and was assigned accession number EF988092. Mary Heidenreich (Warriner) was an undergraduate in the Department of Bacteriology when she conducted this study.
References
An, Z., Mei, B., Yuan, W. and S. A. Leong. 1997a. The distal GATA sequences of the sid1 promoter mediate repression of siderophore production and interact directly
with Urbs1, a GATA family transcription factor. EMBO J. 16:1742 - 1750.
An, Z., Zhao, Q., McEvoy, J., Yuan, W., Markley, J. and S. A. Leong. 1997b. The C-terminal finger domain of Urbs1 is required for iron-mediated regulation of siderophore biosynthesis in Ustilago maydis. Proc. Natl. Acad. Sci. U.S.A. 94:5882-5887.
Gold, S., Bakkeren, G., Davies, J.E., and J.W. Kronstad. 1994. Three selectable markers for transformation of Ustilago maydis. Gene 142:225-230.
Holden, D., J. Wang and S. A. Leong. 1988. DNA-mediated transformation of Ustilago hordei and Ustilago nigra. Physiol. Mol. Pl. Pathol. 33:235-239.
Holliday, R. 1974. Ustilago maydis. In: Handbook of Genetics, Vol. 1 (R. C. King, ed.) Plenum Press, New York. pp. 575–595
Kamper et. al., 2006. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444:97-101.
Leong, S. A. and G. Winkelmann. 1998. Molecular biology of iron transport in fungi. In: Metal Ions in Biological Systems, Vol. 35, Iron Transport and Storage in Microorganisms, Plants and Animals (A. Sigel and H. Sigel, eds.) Marcel Dekker, New York. pp.147-186
Maniatis, T., Fritsch, E. F. and Sambrook, J. 1982. Molecular Cloning: A laboratory Manual. Cold Spring Harbor Laboratory, New York.
Mei, B., Budde, A. D. and S. A. Leong. 1993. sid1, a gene initiating siderophore biosynthesis in Ustilago maydis: Molecular characterization, regulation by iron and role in phytopathogenicity. Proc. Natl. Acad. Sci. U. S. A 90:903-907.
Schendel, F. J., Mueller, E. , Stubbe, J., Shiau, A. and Smith J. M. 1989. Formylglycinamide ribonucleotide synthase from Escherichia coli: Cloning, sequencing, overproduction, isolation, and characterization. Biochemistry 28:2459-2471.
Sherman, F. 1998. An Introduction to the Genetics and Molecular Biology of the Yeast Saccharomyces cerevisiae. Modified from: F. Sherman, Yeast genetics. The Encyclopedia of Molecular Biology and Molecular Medicine, pp. 302-325, Vol. 6. Edited by R. A. Meyers, VCH Pub., Weinheim, Germany,1997. http://dbb.urmc.rochester.edu/labs/sherman_f/yeast/Index.html
Voisard, C., Wang, J., McEvoy, J., Xu, P. and S. A. Leong. 1993. urbs1, a gene regulating siderophore biosynthesis in Ustilago maydis encodes a protein similar to the erythroid transcription factor GATA-1. Mol. Cell. Biol. 13:7091-7100.
Wang, J., Budde, A. and S. A. Leong. 1989. Analysis of ferrichrome biosynthesis in the phytopathogenic fungus Ustilago maydis: Cloning of an ornithine-N5-oxygenase gene. J. Bacteriol. 171:2811-2818.
Yuan, M. W., Gentil, G. D., Budde, A. D. and S. A. Leong. 2001. Characterization of the Ustilago maydis sid2 gene encoding a multidomain peptide synthetase in the ferrichrome biosynthetic gene cluster. J. Bacteriol. 183:4040-4051.