Perspectives on genetic resources at the Fungal Genetics Stock Center.
Kevin McCluskey and Michael Plamann. School of Biological Sciences, University of Missouri- Kansas City.
Fungal Genetics Reports
55:15-17
(Pdf)
The Fungal Genetics Stock Center has been banking and distributing resources for work with genetically characterized fungi since 1960. While most of the collection consists of strains of Neurospora, an NIH model filamentous fungus, the past fifteen years has seen the collection expand to include plant and human pathogenic fungi. The use of the resources in the collection has grown over the last 10 years as well, reflecting both a growth in research using standardized materials as well as the development of new materials through molecular genetic technology. This growth is not limited to newly deposited materials, however, and includes renewed interest in particular classes of strains with characteristics that were not recognized when they were originally deposited. One significant example is the use of strains carrying the osmotic-2 lesion in Neurospora crassa. This, and other utilization trends, underscores the need to provide strong support to the continued and expanded biobanking effort in the US.
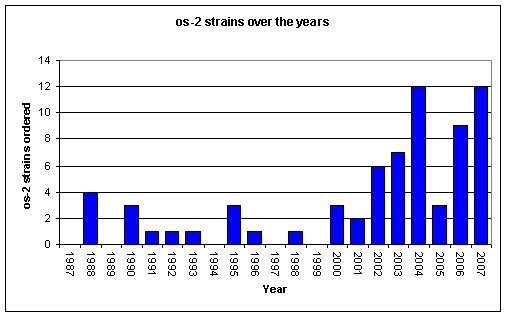
While these modern materials have been popular, there have been numerous occasions where materials that have been in the collection for many years suddenly take on increased importance. One clear example of this is the renewed interest in strains of Neurospora crassa carrying the osmotic-2 lesion. The FGSC holds 66 N. crassa strains carrying mutations leading to sensitivity to high salt concentrations. There are eleven named osmotic loci in Neurospora and os-1 occurs in 38 different strains. The os-2 lesion occurs in only eight strains, the first of which was deposited in 1968. These strains were ordered infrequently through their early years at the FGSC (Figure 1). In 1999, however, Xu and colleagues reported at the 20th Fungal Genetics Conference (Zhang et al., 1999) that strains carrying os-2 were resistant to phenylpyrrole fungicides. This work was later published in Applied and Environmental Microbiology (Shang et al, 2002) and has been cited over 60 times in 5 years.

Figure 1. Orders for strains carrying the os-2 lesion in the last 20 years.
Prior to the 1999 report of the fungicide resistance of os-2 mutants, these strains were used primarily as genetic markers for mapping studies. After this information was presented, requests for strains carrying os-2 jumped over 300%. By way of contrast, strains carrying other osmotic markers have shown only a 29% increase over the same period (Figure 2). These orders came from 32 different investigators in 12 countries.
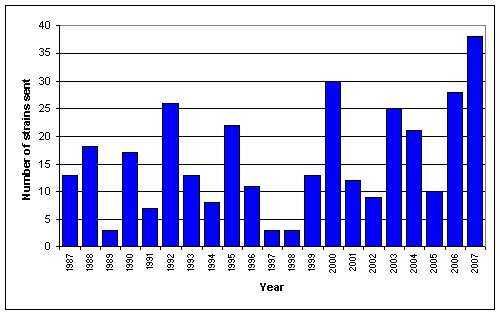
Figure 2. Orders for strains carrying any osmotic lesion over the last 20 years.
The impact of the finding that the original os-2 mutant is resistant to fungicides has spilled over into the usefulness of modern molecular genetic tools. The FGSC is part of an NIH program project to systematically delete every gene in the genome (Dunlap et al., 2007). The most commonly ordered strain from this program is one carrying a knockout of the os-2 gene (Table 1).
|
Mutation |
Number of orders |
|
NCU07024.2 (os-2) |
15 |
|
NCU06182.2 (MAPKK kinase) |
14 |
|
NCU05495.2 (hypothetical) |
13 |
|
NCU03164.2 (hypothetical) |
12 |
|
NCU07221.2 (histidine kinase) |
10 |
|
NCU02057.2 (putative kinase) |
10 |
|
NCU04834.2 (phytochrome) |
8 |
|
NCU02133.2 (sod-1) |
8 |
|
NCU02815.2 (os-1) |
8 |
|
NCU03894.2 (putative kinase) |
8 |
Table 1. Most commonly ordered N. crassa Knock-Out mutants.
While the strains carrying os-2 were used by some investigators prior to the demonstration of their fungicide resistance, they were not among our most popular strains. Between 1987 and 1998, the FGSC distributed over 12,500 strains whereas only 15 os-2 strains were distributed. Had we been required to keep only strains that were among our most well utilized, we might have chosen to 'de-accession' these osmotic mutants. Given the impact of the modern research with them, losing these strains would have been a significant setback to the progress of research into fungicide resistance.
While some existing material in the biological repositories may not show obvious and immediate value, there is no way to predict what their value may be to future researchers. The incremental cost of maintaining additional strains is small (Baker, 2004) and trivial compared to the potential cost of repeating past work or overlooking important opportunities. This example demonstrates the need for continued support for bio-banking activities including those at the FGSC.
The FGSC is supported by award number 0235887 from the National Science Foundation and receives additional funds from award 5P01GM068087-04 from the National Institutes of Health.
References:
Baker, D.D. 2004. Industrially-held microbial diversity: Are culture collections really necessary? In "Microbial Genetic Resources and Biodiscovery." (Kurtboke and Swings, Eds.) World Federation for Culture Collections. pp 261-269.
Dunlap, J.C. Borkovich, K.A., Henn, M., Turner, G., Sachs, M.S., Glass, N.L., McCluskey, K. Plamann, M., Galagan, J.E., Birren, B. W., Weiss, R.L., Townsend, J.P., Loros, J.J. Nelson, M.A., Lambreghts, R., Colot, H.V. Park, G., Collopy, P., Ringelberg, C., Crew, C. Litvinkova, L., DeCaprio, D., Hood, H.M., Curillo, S., Shi, M., Crawford, M., Koerhsen, M., Montgomery, P., Larson, L., Pearson, M., Kasuga T., Tian, C., Batürkmen, M., Altamirano, L., and Xu. J. 2007. Enabling a community to dissect an organism— Overview of the Neurospora Functional Genomics Project. Adv. Genet. 57: 49-96.
McCluskey, K. 2003. The Fungal Genetics Stock Center: from Molds to Molecules.. In "Advances in Applied Microbiology Volume 52." (Laskin and Bennett, Eds.) Elsevier USA. pp 246-262.
Zhang, Y., R. Lamm, C. Pillonel, Xu, J.R. and S. Lam. 1999. The hyper-osmotic stress response pathway of Neurospora crassa is the target of phenylpyrrole fungicides. Fungal Genet. Newsl.46(S):122.
Zhang, Y., R. Lamm, C. Pillonel, S. Lam, and J. R. Xu. 2002. Osmoregulation and fungicide resistance: the Neurospora crassa os-2 gene encodes a HOG1 mitogen-activated protein kinase homologue. Appl. Environ. Microbiol. 68:. 532-538.