Visualization of peroxisomes via SKL-tagged DsRed protein in Sordaria macrospora
Skander Elleuche and Stefanie Pöggeler*
Georg-August-University of Göttingen, Institute of Microbiology and Genetics, Department of Genetics of Eukaryotic Microorganisms. *corresponding author, e-mail: spoegge@gwdg.de;
Fungal Genetics Reports 55:9-12
We report the utilization of Discosoma sp. red fluorescent protein DsRed to visualize peroxisomes in the filamentous ascomycete Sordaria macrospora. To achieve labeling of peroxisomes, DsRed was fused to a serine-lysine-leucine tag (SKL). Expression of the DsRed-SKL fusion gene under the control of the Aspergillus nidulans gpd-promoter led to protein import of DsRed into peroxisomes. In this study, we describe our results concerning the construction as well as the application of vector pDsRed-SKL.
The green fluorescent protein GFP from the jellyfish Aequorea victoria and other fluorescent proteins have become standard tools for the investigation of a variety of cellular activities (Prasher 2005). Proteins with different spectral properties and the fusion of fluorescent proteins with specific organelle-targeting signals are preconditions to analyze the sub-cellular localization of a given protein. In filamentous fungi, beside GFP and its variants, engineered derivatives of the Discosoma red fluorescent protein DsRed have been applied for dual-color experiments (Freitag and Selker 2005; Mikkelsen et al. 2003; Pöggeler et al. 2003; Toewes et al. 2004).
A standard procedure to localize a given protein within a fungal cell is the expression of its GFP-tagged version and its co-localization with either stained cell compartments or fluorescent tagged marker proteins targeted to defined organelles. Here, we demonstrated the peroxisomal localization of an SKL-tagged DsRed protein in the homothallic ascomycete S. macrospora, which is a close relative to the heterothallic fungus Neurospora crassa and a useful model organism for fruiting-body development (Kück and Pöggeler 2005). Previously, peroxisome-targeting via fusion of GFP with a C-terminal SKL tripeptide, a peroxisomal targeting sequence 1 (PTS1) signal for peroxisomal matrix import, has been reported for Podospora anserina (Ruprich-Robert et al. 2002). The P. anserina plasmid pGPD::GFP-SKL was also shown to be suitable for labeling of peroxisomes in S. macrospora (Engh et al. 2007).
Here, we constructed a DsRed-SKL plasmid on which the DsRed-SKL gene is driven by the constitutive gpd promoter of Aspergillus nidulans. This plasmid enables the co-localization with GFP-tagged proteins.
The coding sequence of the DsRed-gene was amplified with primer pair DsRed_for (5’-CATCACCATGGCCTCCTCCGAGG; NcoI restriction underlined)/DsRedSKL_rev GGATCCTTAGAGCTTGCTCAGGAACAGGTGGTGGCGGCCC; BamHI restriction site underlined; SKL coding sequence as well as a new inserted STOP-codon in bold italics). The base-triplets encoding the SKL-motif were adapted to the codon usage of S. macrospora (Pöggeler 1997). We used plasmid pRHN1 as a template carrying the DsRed gene (DsRed-Express; BD Biosciences Clontech, Heidelberg, Germany) and the nourseothricin resistance gene nat1 as a selective marker (Janus et al. 2007). The obtained 700-bp DsRed-SKL amplicon was subcloned into vector pDrive (Qiagen, Hilden, Germany). After sequence verification (MWG-Biotech Customer Service, Ebersberg, Germany), the plasmid was digested with BamHI and NcoI. Subsequently, the DsRed gene of plasmid pRHN1 was replaced with DsRed-SKL. The resulting plasmid was termed pDsRed-SKL (Fig. 1).
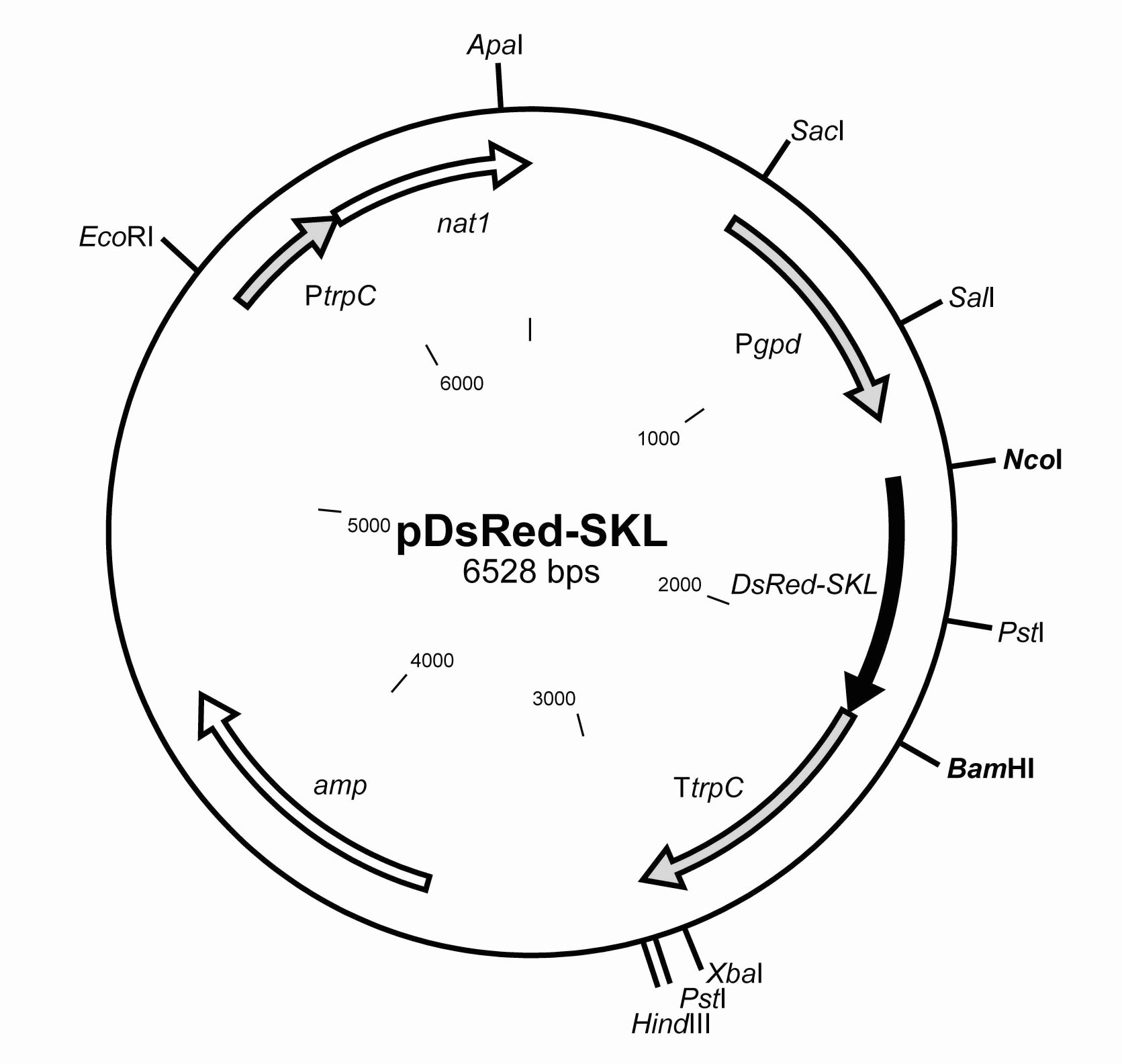
Figure 1: Partial restriction map of vector pDsRed-SKL. The ampicillin resistance cassette is indicated as amp and can be used for selection of Escherichia coli transformants. The nourseothricin resistance gene nat1 (white arrow) under control of the A. nidulans trpC promoter can be used for selection of fungal transformants. The DsRed-SKL (black arrow) is regulated by the gpd promoter and the trpC terminator of A. nidulans. All promoter and terminator sequences are indicated as grey arrows.
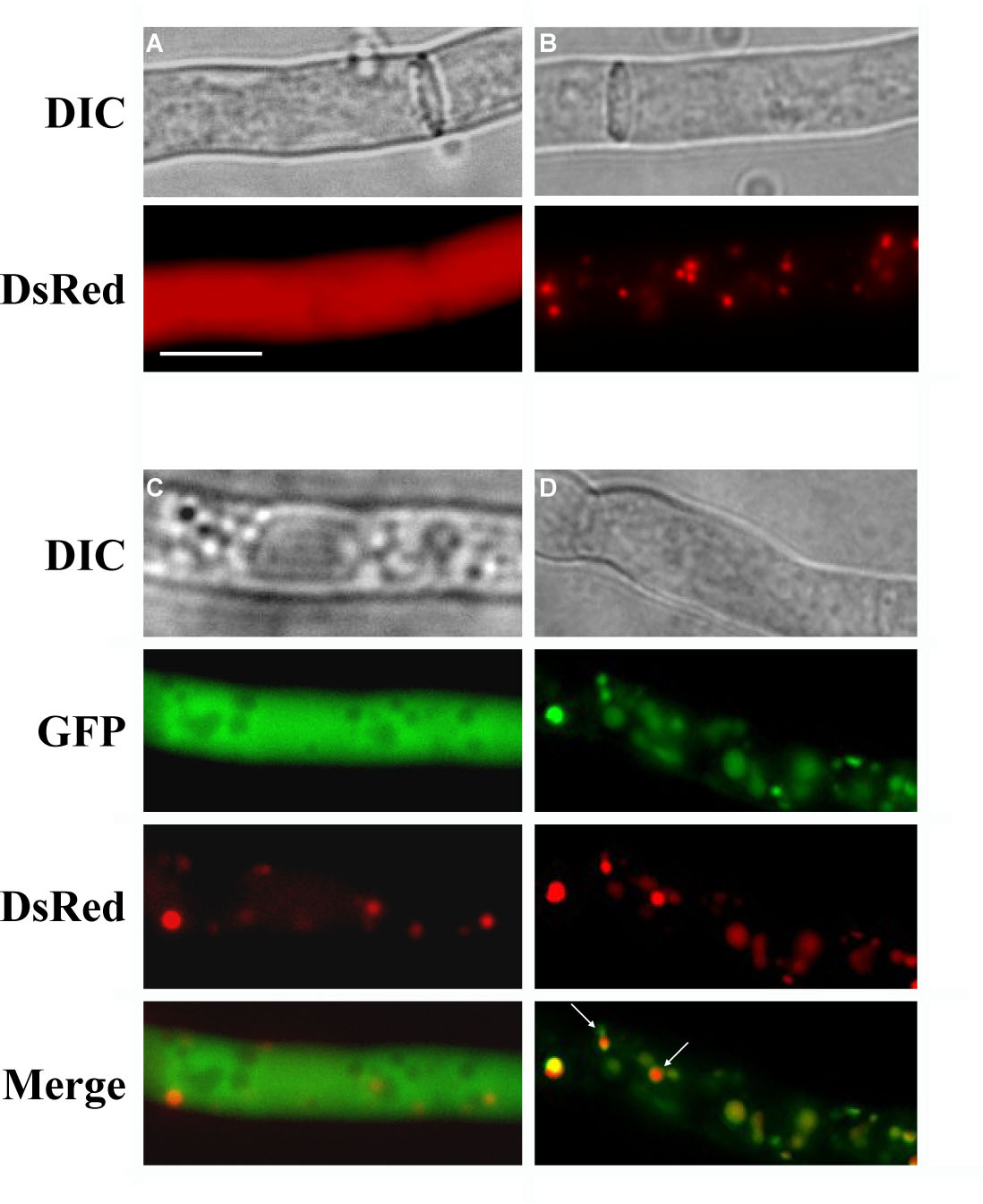
To verify the functionality of the DsRed encoded by pDsRed-SKL, we transformed a S. macrospora wildtype strain (Culture collection, Department for General and Molecular Botany, Ruhr-University of Bochum) either with plasmid pRHN1 or pDsRed-SKL (Fig. 2A and 2B).

Figure 2. Expression of cytosolic and peroxisomal targeted DsRed and GFP in S. macrospora. (A) Hyphae of a strain transformed with pRHN1. (B) Hyphae of a strain transformed with pDsRed-SKL. Expression of DsRed-SKL leads to DsRed import into peroxisomes, visible as red dots in the cytoplasm. (C) Co-transformation with pIG1783 and pDsRed-SKL results in green fluorescent hyphae with red colored peroxisomes. (D) Parallel expression of SKL-tagged GFP and SKL-tagged DsRed. Both tagged proteins are not imported into the same exact organelle at the same rate, resulting in green or red dots after merging (see white arrows). Scale bar, 10 µm. A color reproduction of the figure is available with the on-line version of this article (http://www.fgsc.net/).
Transformants were selected on CMS-medium (0.15 % (w/v) KH2PO4; 0.05 % (w/v) KCl; 0.05 % (w/v) MgSO4; 0.37 % (w/v) NH4Cl; 1 % (w/v) glucose; 0.2 % (w/v) tryptone; 0.2 % (w/v) yeast extract; 10.8 % (w/v) saccharose; trace metals [10 mg/l ZnSO4; 10 mg/l Fe(II)Cl2; 10 mg/l MnCl2]; 2 % (w/v) agar; pH 6.5) containing nourseothricin (50 µg/ml). To demonstrate the functionality of the DsRed-SKL protein in dual-color experiments, we combined the red peroxisomal label (pDsRed-SKL) with cytosolic GFP encoded by plasmid pIG1783 (Pöggeler et al. 2003; Fig. 2C) or the green peroxisomal label GFP-SKL (pGPD::GFP-SKL; Ruprich-Robert et al. 2002; Fig. 2D). Co-transformants were selected either on CM-medium containing nourseothricin (50 µg/ml) and hygromycin (110 U/ml) (pDsRed-SKL and pIG1783) or on medium containing solely nourseothricin (50 µg/ml) (pDsRed-SKL and pGPD::GFP-SKL), since pGPD::GFP-SKL carries a phleomycin-resistance cassette, which is not selectable in S. macrospora.
Many yeast species and filamentous ascomycetes including Acremonium chrysogenum, Neurospora crassa and A. nidulans are sensitive to nourseothricin (Kück and Hoff 2006, Braus and Seiler, pers. communication). The antibiotic nourseothricin is commercially available (Werner BioAgents, Jena, Germany) and similarly expensive as hygromycin. Transformants and co-transformants were then inoculated on microscope slides overlaid with a thin layer of solid SWG minimal medium (0.4 % (w/v) arginine; 0.1 % (v/v) biotin; 2 % (w/v) glucose; 1x Westergaard (Westergaard and Mitchell 1947); 1.5 % (w/v) agar; pH 6.5) containing either nourseothricin or hygromycin and nourseothricin. After incubation for three days at 27 °C, fluorescent proteins were visualized with appropriate filter combinations (Chroma filter set 49002, exciter ET470/40x, emitter ET525/50m and beamsplitter T495LP; Chroma filter set 49005 for green fluorescence, exciter ET545/30x, emitter ET620/60m and beamsplitter T570LP for red fluorescence) using an AxioImager M1 microscope (Zeiss, Jena, Germany) and an X-cite 120 PC lamp (EXFO, Ontario, Canada). Images were captured with a Photometrics CoolSNAP HQ2 camera (Roper Scientific, Photometrics, Tucson, Arizona). Recorded images were edited with MetaMorph (VisitronSystems, Puchheim, Germany) and Adobe Photoshop CS2.
As expected, fluorescence appeared uniformly distributed throughout the cytoplasm of the hyphae in transformants carrying pRHN1 while transformation with pDsRed-SKL led to a punctate fluorescent pattern (Fig. 2A, B). An SKL-tagged DsRed used for peroxisomal studies has recently been described for the basidiomycete Cryptococcus neoformans. Similar to our results, a C. neoformans DsRED-SKL strain exhibited a punctate peroxisomal fluorescence (Idnurm et al. 2007).
Furthermore, we tested the fluorescence efficiency of DsRed-SKL in a cytoplasmic GFP-background by co-transformation of pDsRed-SKL and pIG1783 under nourseothricin/hygromycin double-selection (Fig. 2C). Transformants carrying both plasmids displayed a well-defined punctate red fluorescent pattern clearly visible against the cytosolic background of GFP. To confirm that DsRed-SKL actually localizes to peroxisomes, we accomplished co-localization experiments with GFP-SKL. For this purpose, we analyzed 38 nourseothricin-resistant transformants from a pDsRed-SKL/pGPD::GFP-SKL co-transformantion experiment. More then 55 % of the investigated transformants (21 transformants) exhibited DsRed as well as GFP fluorescence, while the remaining transformants either emitted only the red fluorescence od DsRed-SKL or no fluorescence. As shown in Fig. 2D, the DsRed-SKL fusion produced a fluorescence pattern, which coincides with the fluorescence pattern of the GFP-SKL. Thus the DsRed-SKL fusion construct is targeted to peroxisomes. However, because of rapid peroxisome movement in the cytosol, signals did not align perfectly in all cases (Fig. 2D).
In conclusion, expression of pDsRed-SKL leads to the import of DsRed into peroxisomes and results in the detection of fluorescence signals after excitation with green light. Vector pDsRed-SKL leads to high-level expression of the DsRed-SKL gene and the import of DsRed into peroxisomes.
Plasmid pDsRed-SKL is consequently a valuable tool for analyzing the sub-cellular localization, particularly the peroxisomal localization, of GFP-tagged proteins by means of dual-color experiments. It could be also used for the analysis of mutants affected in peroxisome biogenesis. The vector was primarily constructed for S. macrospora; however, it should prove very effective in a wide variety of filamentous ascomycetes in which transformation is based on nourseothricin resistance.
Acknowledgements:
We gratefully acknowledge Prof. Ulrich Kück (Department of General and Molecular Botany, Ruhr-University of Bochum, Germany) for kindly providing DsRed vector pRHN1 and we wish to thank Dr. Nicole Nolting for critically reading the manuscript.
References:
Engh, I., Würtz, C., Witzel-Schlömp, K., Zhang, H.Y., Hoff, B., Nowrousian, M., Rottensteiner, H., Kück, U., 2007. The WW Domain Protein PRO40 is required for fungal fertility and associates with woronin bodies. Eukaryot. Cell 6: 831-843
Freitag, M., and Selker, E.U., 2005. Expression and Visualization of Red Fluorescent Protein (RFP) in Neurospora crassa. Fungal Genet. Newslett. 52: 14-17
Idnurm, A., Giles, S.S., Perfect, J.R., Heitman, J., 2007. Peroxisome function regulates growth on glucose in the basidiomycete fungus Cryptococcus neoformans. Eukaryot. Cell 6: 60-72
Janus, D., Hoff, B., Hofmann, E., Kück, U., 2007. An efficient fungal RNA-silencing system using the DsRed reporter gene. Appl. Environ. Microbiol. 73: 962-970
Kück, U., and Hoff, B., 2006. Application of the nourseothricin acetyltransferase gene (nat1) as dominant marker for the transformation of filamentous fungi. Fungal Genet. Newslett. 53: 9-11
Kück, U., and Pöggeler, S., Sordaria macrospora, 2005. In Production of Recombinant Proteins. Novel Microbial and Eucaryotic Expression Systems., in Gellisen, G., (Ed.), WILEY-VCH, Weinheim, pp. 215-231
Mikkelsen, L., Sarrocco, S., Lubeck, M., Jensen, D.F., 2003. Expression of the red fluorescent protein DsRed-Express in filamentous ascomycete fungi. FEMS Microbiol. Lett. 223: 135-139
Pöggeler, S., 1997. Sequence characteristics within nuclear genes from Sordaria macrospora. Fungal Genet. Newslett. 44: 41-44
Pöggeler, S., Masloff, S., Hoff, B., Mayrhofer, S., Kück, U., 2003. Versatile EGFP reporter plasmids for cellular localization of recombinant gene products in filamentous fungi. Curr. Genet. 43: 54-61
Prasher, D.C., 1995. Using GFP to see the light. Trends Genet. 11: 320-323
Ruprich-Robert, G., Berteaux-Lecellier, V., Zickler, D., Panvier-Adoutte, A., Picard, M., 2002. Identification of six loci in which mutations partially restore peroxisome biogenesis and/or alleviate the metabolic defect of pex2 mutants in podospora. Genetics 161: 1089-1099
Toews, M.W., Warmbold, J., Konzack, S., Rischitor, P., Veith, D., Vienken, K., Vinuesa, C., Wei, H., Fischer, R. 2004. Establishment of mRFP1 as fluorescent marker in Aspergillus nidulans and construction of expression vectors for high-throughput protein tagging using recombination in vitro (GATEWAY). Curr. Genet. 45: 383-389
Westergaard, M., and Mitchell, H.K. 1947. Neurospora V. A synthetic medium favoring sexual reproduction. Am. J. Bot. 34: 573-577