A versatile set of Lifeact-RFP
expression plasmids for live-cell imaging of F-actin
in filamentous fungi
Alexander Lichius
and Nick D. Read
Fungal Cell Biology Group, Institute of Cell Biology, University of Edinburgh,
Rutherford Building, Edinburgh EH9 3JR, UK;
Alex@fungalcell.org
Fungal Genetics Reports 57:8-14
Here we report the construction and application of a range of expression plasmids designed to facilitate live-cell imaging of F-actin dynamics in filamentous fungi simultaneously with other, preferably GFP-tagged fusion proteins. Pros and cons of the use of three different red fluorescent proteins (RFPs), two different promoters and three different selection markers are addressed.
Live-cell imaging of F-actin dynamics is possible in a wide range of eukaryotes.
Lifeact is a 17 aa peptide derived from the actin-binding protein 140 (Abp140) of S. cerevisiae which specifically binds to filamentous actin (F-actin) (Riedl et al., 2008). Functionality of green fluorescent Lifeact reporters (Lifeact-GFP) for the visualisation of F-actin structures in living cells has so far been documented for three of the four eukaryotic phyla, including yeasts and filamentous fungi (Berepiki et al., 2010; Böhmer et al., 2009; Coffman et al., 2009; Delgado-Álvarez et al., 2010; Riedl et al., 2008), plants (Era et al., 2009; Smertenko et al., 2010; Vidali et al., 2009), and mammals (Estecha et al., 2009; Riedl et al., 2008; Riedl et al., 2010). The application of Lifeact-GFP in filamentous fungi has been pioneered in the ascomycete Neurospora crassa (Berepiki et al., 2010; Delgado-Álvarez et al., 2010), and is currently being adopted for the use in numerous other fungal species. In basidiomycetes, however, labelling properties of Lifeact appear to be restricted to the visualization of septal rings, as F-actin cables and patches have so far not been explicitly reported (Böhmer et al., 2009). Lifeact-GFP reporters work equally well in fungi that use the CUG codon for serine and not leucine, such as Candida sp. (Kawaguchi et al., 1989), once they have been codon corrected, (E. Epp, McGill Univ. Montreal, pers.comm.).
Why RFP and which one?
The most widely used fluorescent
reporter for live-cell imaging analyses of protein dynamics in filamentous
fungi is GFP. To allow simultaneous observation of F-actin with any other GFP-tagged
protein, the development of a Lifeact probe (Riedl et al.,
2008) labelled with a different color was desired. As
green and red emission signals can be spectrally well separated, and
fluorescence microscopes with the corresponding excitation and emission
detection settings are widely available, we aimed to generate a functional Lifeact-RFP reporter construct. The first red fluorescent Lifeact probe that we adopted for the filamentous model
fungus Neurospora crassa was Lifeact-tdTomato (Roca et al., 2010). Although Lifeact-tdTomato reliably labelled
F-actin patches, cables and septal
rings (Figure 1), this reporter occasionally produced additional ring-shaped
structures (~1-2 µm in diameter, Figures 1B and C),
which we suggest may be formed through spontaneous self-assembly of F-actin, destabilized through the interaction with the Lifeact-tdTomato construct, and native septins.
Evidence for this comes from previous studies indicating that due to sterical hindrance, mRFP and tdTomato fusion constructs failed to correctly localize
microtubules and connexin (Shaner
et al., 2004; Shaner et al., 2008). This not well
understood property of some RFPs might have
potentiated mislocalization artefacts
of the overexpressed Lifeact-tdTomato
fusion construct leading to the formation of these cytoplasmic
rings from destabilized F-actin. Interestingly, similar
structures have been reported to form upon Latrunculin
A treatment in mammalian cells (Kinoshita et al., 2002) and haploid cells of Ustilago maydis (Böhmer et al.,
2009). Appearance of these rings in N.
crassa was resolved by exchanging tdTomato for the
monomeric red fluorescent protein TagRFP
(Shaner et al.,
2008).
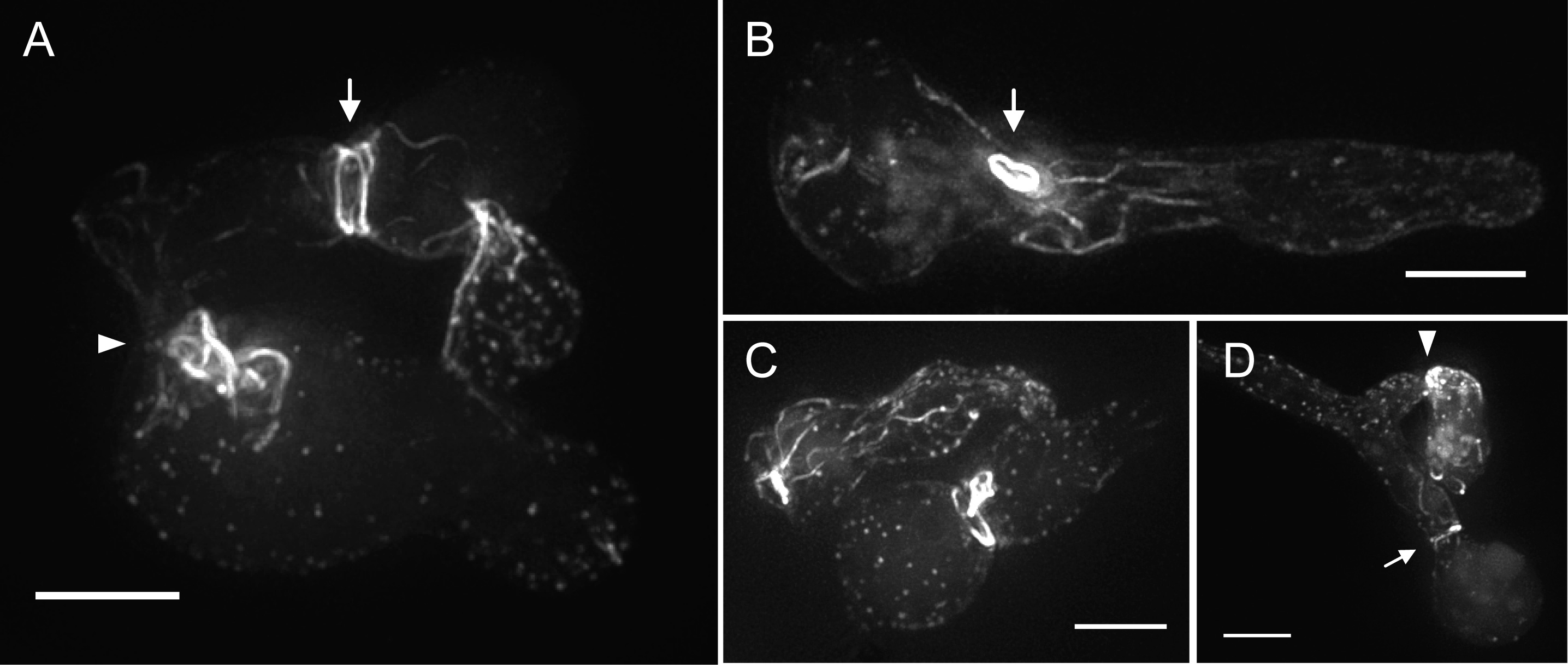
Figure 1. Lifeact-RFPs
labelled F-actin cables,
patches and rings. (A) Two fused wt cells expressing Lifeact-TagRFP.
F-actin cables stretched throughout the spore body of
the upper cell, whilst actin patches were distributed
over the surface of the bottom cell. Dense arrays of actin
cables were localized at the cell fusion site (arrowhead), and condensed into
an actin ring prior to septum formation (arrow). (B
to D) Examples of wt conidia expressing Lifeact-tdTomato.
(B) Conidial germling with actin
cables aligned with the long axis of the germ tube. An extremely bright
fluorescent ring (arrow) in the cytoplasm is obvious but not associated with
any known cellular structure. Similar rings have been described in other model
systems and are likely the product of spontaneous self-assembly of the Lifeact-tdTomato construct with native septins
(see text for details). Also vacuolar accumulation of the fusion construct
created minor artefactual background fluorescence.
(C) Two fusing germlings with bright fluorescent
rings at different localizations within the cells. (D) Conidia in the process
of CAT homing (Read et al., 2009), showing intense accumulation of actin patches and cables in the tip of the smaller germling (arrowhead). Formation of a cortical actin ring at the spore neck is again indicated with an
arrow. All images show maximum intensity projection of deconvolved
z-stacks. Scale bars, 5 µm.
To ensure an equivalent high level of fluorescence regardless of its use as a N- or C-terminal fusion tag the original TagRFP was modified through the addition of GFP-like N- and C- terminal residues (VSKGEE and LNGMDELYK, respectively), thereby improving its utility for 4D live-cell imaging (Berepiki et al., 2010; Shaner et al., 2004; Shaner et al., 2008). Finally, the photostability of this optimized TagRFP version was increased through site-directed mutagenesis (SDM) of S162T, using the oligonucleotides TagRFP_S162T_fw 5’-CCTGGAAGGCAGAACCGACATGGCCCTGAAGC-3’ and its reverse complement TagRFP_S162T_rv 5’-GCTTCAGGGCCATGTCGGTTCTGCCTTCCAGG-3’ (the mismatching nucleotide conferring S162T exchange is underlined; the affected triplet is in bold and italicised). The resulting TagRFP-T displayed a 10-times lower average decay constant (k) compared to TagRFP in living cells of N. crassa (Figure 2), confirming its status as the most photostable monomeric FP so far described (Shaner et al., 2008). Furthermore, TagRFP-T, in contrast to TagRFP, although showing a small, ~17% decrease in brightness, shows virtually no emission in the green part of the spectrum (Shaner et al., 2008). Therefore, Lifeact-TagRFP-T is ideally suited for co-imaging with GFP-tagged proteins, for example BML-sGFP which reliably labels β-tubulin in microtubules (Berepiki et al., 2010; Freitag et al., 2004).
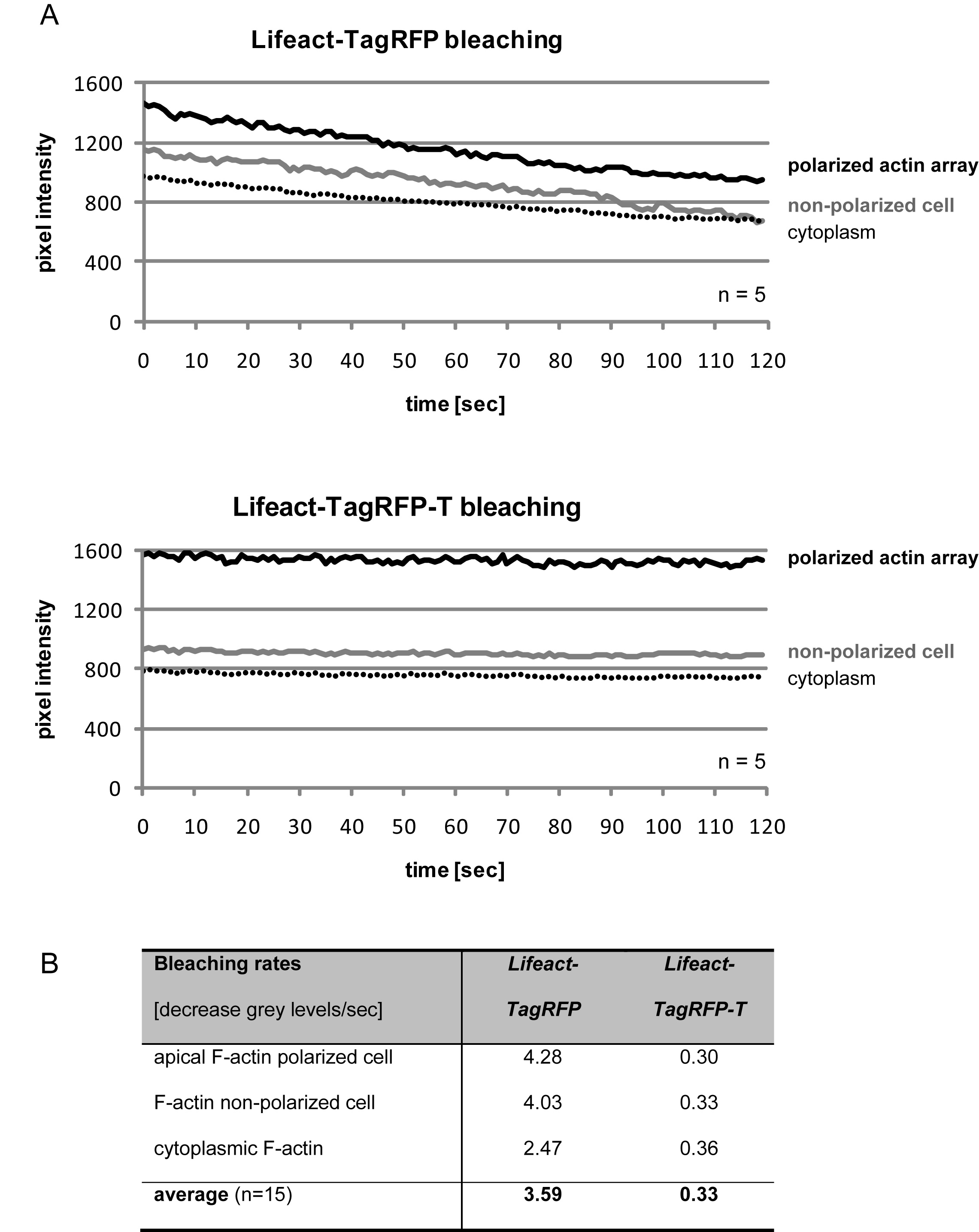
Figure 2. Lifeact-TagRFP-T is on average 10x more photostable than Lifeact-TagRFP. (A) Cells expressing Lifeact-TagRFP or Lifeact-TagRFP-T were imaged continuously over a period of 2 min with 1 frame/sec in one focal plane and 0.5 sec exposure using 60% illumination intensity from a 550 nm LED excitation light source. Bleaching rates were measured as the decrease in grey levels per second in 20 µm² regions of interest drawn up within three different intracellular areas containing different amounts of F-actin as judged by initial Lifeact-FP signal intensity: cortical F-actin at germ tube tip, cortical F-actin in non-polarized cell, and cytoplasmic F-actin in the cell centre of a polarized growing germling. Graphs represent the means of measurements from five individual cells. (B) On average, Lifeact-TagRFP lost 3.6 grey levels/sec, whereas Lifeact-TagRFP-T only bleached with 0.33 grey levels/sec, corresponding to average decay constants (k) of 0.003 ± 0.0007 and 0.0004 ± 0.0002, respectively
Although no mislocalization
of a range of different TagRFP/RFP-T fusion
constructs has been reported in mammalian cells (Shaner et al., 2008) - which we have confirmed
for a number of other fusion constructs in N.
crassa, including MAP kinases (OS-2-TagRFP-T) (A. Lichius and N. D.
Read, in prep.) and Cdc42-Rac1-Interactive-Binding
(CRIB-TagRFP-T) reporters (A. Lichius,
A. Goryachev, M. D. Fricker,
B. Obara and N. D. Read, in prep.) - C-terminal
addition of TagRFP/RFP-T did occasionally result in
increased vacuolar accumulation of the fluorescence signal in N. crassa. This did not occur with
the corresponding GFP-tagged versions of these
constructs, suggesting this phenomenon might be associated with the increased
stability of TagRFP/RFP-T at low pH compared to other
FPs (pKa: TagRFP < TagRFP-T < tdTomato < EGFP, Shaner et al.,
2008). In other words, the amount of vacuolar accumulation associated with the
normal turn-over of the fluorescent reporter proteins might not be different
between GFP and TagRFP/RFP-T
tagged constructs, but due to the increased pH stability of RFPs
their degradation progresses much slower and thus they are more readily
detectable. This unwanted side effect can hamper live-cell imaging
significantly as it decreases the signal-to-noise ratio. It can be minimized by
selecting transformants with low plasmid integration
number, and through the use of a weaker promoter driving reporter expression
(see below). The low binding affinity of the Lifeact
peptide to F-actin and its high on/off rate (Riedl et al.,
2008) are the likely reasons why these unwanted secondary effects involving (1)
aggregation of the reporter construct with endogenous proteins and (2)
pronounced vacuolar accumulation, do generally not occur with the Lifeact-TagRFP/RFP-T probes. With our current level of
understanding, for co-localization studies it seems advisable to use the Lifeact peptide with a TagRFP-T
label and tag the other protein under investigation with GFP.
This will ensure optimal performance of both fluorescent fusion constructs.
Which promoter provides stable expression throughout the fungal lifecycle?
So far, the most commonly used promoter for ectopic expression of FP fusion constructs in Neurospora crassa is Pccg-1 (Freitag et al., 2004; McNally and Free, 1988). Pccg-1 regulates expression of the clock-controlled gene 1 (ccg-1) in a very complex manner, involving at least circadian clock control, light control, metabolic control, and very probably developmental regulation (Arpaia et al., 1995; Loros et al., 1989). Overall, this results in very strong but not necessarily consistent expression throughout development. Heterologous expression under its regulation is induced by light (Arpaia et al., 1995), and highest in conidia and aerial hyphae, including enveloping hyphae during protoperithecial development (A. Lichius and K.M. Lord, C. E. Jeffree and N. D. Read, in prep.). In conidial germlings we consistently observed a noticeable decrease in expression and thus recordable signal intensity after 6-7 h of incubation (about 5-6 h after cell symmetry breaking of the germinating conidium). We also found that Pccg-1 did not allow satisfactory visualization of Lifeact-FPs in mature vegetative hyphae (Berepiki et al., 2010). To fully overcome these problems and guarantee stable fluorescent signal levels throughout the whole life cycle of the fungus, the promoter of the translation elongation factor-1 (locus NCU02003.4) (Ptef-1) of Neurospora crassa was alternatively employed (Berepiki et al., 2010). In summary, for studies during the first 6-7 h of conidial development, Pccg-1 is a very well suited promoter for N. crassa, whereas expression under its control appears less consistent during the transition from the germling phase into mature hyphal growth. The use of Ptef-1, which gives overall weaker but more consistent expression throughout all developmental stages of N. crassa is an alternative worth considering.
Which selection marker allows transformation of Neurospora gene deletion strains?
An interesting question to address is how deletion of specific genes, e.g. components of the cell polarity machinery, influence F-actin architecture and functionality. To facilitate utilization of the Lifeact-RFP expression plasmids in all gene deletion strains generated within the Neurospora Genome Project (http://www.dartmouth.edu/~neurosporagenome/), it was imperative to employ a selection marker other than hygromycin B, as the hygromycin B resistance gene hph has been used as a central part of the gene knock-out cassette. The two resistance genes exploited in this study were bar, conferring resistance towards ignite (also known as phosphinothricin or basta) (Pall and Brunelli, 1994), and nat1, conferring resistance towards nourseothricin (Kück and Hoff, 2006). Ignite resistance has been employed for the generation of ∆mus-51 and ∆mus-52 gene deletion host strains (Colot et al., 2006), thus it cannot be used for the transformation of knock-out strains in which the ∆mus deletion cassette has not been removed through backcrossing to the wild type. Furthermore, ignite selection in Neurospora is relatively leaky compared to other commonly used selection markers, especially when using the potentially impure chemical extracted from the commercially available “Harvest” herbicide (Bayer Crop Sciences Ltd., Monheim, Germany) (Hays and Selker, 2000; Metzenberger et al., 2000). Therefore, pALx-Lifeact transformant selection with nourseothricin was the preferred option, as it is much more stringent than ignite per se and has no reported problems in conferring cross-resistance to other selection markers, such as hygromycin B (Kück and Hoff, 2006). Nourseothricin has been successfully used for the transformation of several yeast species including Saccharomyces cerevisiae (Goldstein and McCusker, 1999), Schizosaccharomyces pombe (Hentges et al., 2005), Candida albicans (Shen et al., 2005) and Cryptococcus neoformans (McDade and Cox, 2001), and more recently in an increasing number of filamentous fungal species, including Acremonium chrysogenum and Sordaria macrospora (Kück and Hoff, 2006), as well as Neurospora crassa (Maerz et al., 2009; Maerz et al., 2008).
Construction of pALx-Lifeact expression vectors.
pAL2-Lifeact, the first vector generated within this project, was assembled by blunt-end ligation of the Lifeact-tdTomato coding sequence amplified from pJT580a into an EcoRI linearized backbone of pBARGRG-1 (Pall and Brunelli, 1994; Roca et al., 2010). pAL3-Lifeact was assembled using InFusion PCR cloning (http://www.clontech.com) to recombine the insert encoding for Lifeact-TagRFP amplified from pJT603f (Jens Tilsner, University of Edinburgh) with the EcoRV/BamHI linarized pBARGRG-1 in a ligation-independent step (Berepiki et al., 2010). Exchange of the ignite resistance cassette (PtrpC::bar) for the nourseothricin resistance cassette (PtrpC::nat1) was accomplished using the same technique to recombine the PtrpC::nat1 insert amplified from pG-Nat1 (Kück and Hoff, 2006) with SpeI/XbaI linearized pAL3-Lifeact, resulting in pAL4-Lifeact. TagRFP in pAL3-Lifeact and pAL4-Lifeact were modified to TagRFP-T by SDM as described above to yield pAL5-Lifeact and pAL6-Lifeact, respectively. Promoter replacement of Pccg-1 with Ptef-1 was performed again using InFusion PCR cloning, but this time integrating the Ptef-1 insert amplified from pTEFsG-2 (Berepiki et al., 2010) into BglII/BamHI linearized pAL4-Lifeact to generate pAL10-Lifeact. In a final SDM step, pAL10-Lifeact was modified to yield pAL12-Lifeact, which expresses Lifeact-TagRFP-T under control of the Ptef-1 promoter, and is selectable with nourseothricin. Table 1 provides an overview of all the plasmids generated; Figure 3 shows examples of the physical vector maps. The sequence of the 17 aa Lifeact reporter (GGVADLIKKFESISKEE) is the same in all constructs and identical to the originally published peptide derived from Abp140 of S. cerevisiae (Riedl et al., 2008). Tables 2 and 3 show primers used for InFusion PCR cloning and sequencing of the recombined vectors, respectively.
Table 1. List of all pALx-Lifeact expression plasmids currently deposited at the FGSC.
|
plasmid |
promoter |
reporter peptide |
fluorescent protein |
terminator |
selection gene/marker |
FGSC # |
|
pAL2-Lifeact |
Pccg-1 |
Lifeact |
tdTomato |
TtrpC |
bar/ignite |
756 |
|
pAL3-Lifeact |
Pccg-1 |
Lifeact |
TagRFP |
TtrpC |
bar/ignite |
757 |
|
pAL4-Lifeact |
Pccg-1 |
Lifeact |
TagRFP |
TtrpC |
nat1/nourseothricin |
758 |
|
pAL5-Lifeact |
Pccg-1 |
Lifeact |
TagRFP-T |
TtrpC |
bar/ignite |
759 |
|
pAL6-Lifeact |
Pccg-1 |
Lifeact |
TagRFP-T |
TtrpC |
nat1/nourseothricin |
760 |
|
pAL10-Lifeact |
Ptef-1 |
Lifeact |
TagRFP |
TtrpC |
nat1/nourseothricin |
761 |
|
pAL12-Lifeact |
Ptef-1 |
Lifeact |
TagRFP-T |
TtrpC |
nat1/nourseothricin |
762 |
Table 2. List of InFusion PCR cloning primers. 15-18 nt overhangs are italicised.
|
name |
5’-3’ sequence |
|
LA_if_BamHI_fw |
TTTCCTCGACGGATCCATGGGTGTCGCAGATTTGATCAAGA |
|
TagRFP_if_EcoRV_rv |
ATCGATAAGCTTGATATCTTACTTGTACAGCTCGTCCATGCCA |
|
PtrpC‐nat1_if_SpeI_fw |
GTTTAGATCCACTAGTCAACTGATATTGAAGGAGCATTTTTTGG |
|
nat1_if_XbaI_rv |
CAGAATGTGCTCTAGATCAGGGGCAGGGCATGCTCA |
|
Ptef-1_if_BglII_fw |
GCGAGGCAGAGATCTCCGTGACCACTGAACTACACTAGTCTAGAG |
|
Ptef-1_if_BamHI_LA_rv |
GACACCCATGGATCCGATATCTTTGACGGTTGATGTGCTG |
Table 3. List of sequencing primers.
|
name |
5’-3’ sequence |
binding site |
|
pAL_seq_fw |
GATTGCCGATTAAGGGAACCAAATG |
end of Pccg-1 |
|
pAL_seq_rev |
AACACCATTTGTCTCAACTCCGGAG |
downstream of FP end |
|
PtrpC_seq_fw |
AGCTAGCTTGGCTGCAGGTC |
upstream of PtrpC start |
|
nat1_seq_rv |
GGGTTATTGTCTCATGAGCGGA |
downstream of nat1 end |
|
Tef1_seq_fw |
GGTGCTTCACAACTCAGCAAATTCTC |
downstream of Ptef-1 start |
|
TagRFP_seq_rv |
CATGTGAAGCCCTCAGGGAA |
inside TagRFP/TagRFP-T |
Lifeact-RFP reporters work in a range of ascomycete fungi.
Besides Neurospora crassa (Roca et al., 2010) and this report), red fluorescent Lifeact reporters have already been successfully expressed and visualized in other ascomycetes, including Sordaria macrospora (S. Poeggeler, Univ. Goettingen, pers. comm.), Magnaporthe oryzae (N.J. Talbot, Univ. Exeter, pers. comm.) and Aspergillus nidulans (B.D. Shaw, Texas A&M Univ., pers. comm.); either by direct or co-transformation of pALx-TagRFP/RFP-T vectors. However, severe overexpression of Lifeact-FP reporters can lead to significant impairment of cell development and colony morphology, and is a problem that currently needs to be resolved for some fungal models, such as Ashbya gossypii (A. Gladfelter, Dartmouth College, pers. comm.). Once suitable expression systems have been identified for those species, the Lifeact-RFP reporters described here should assist and facilitate further and comparative studies of F-actin dynamics in a broad range of filamentous fungi.
All plasmids and a selection of N. crassa strains co-/expressing the various Lifeact-FP reporters are available from the Fungal Genetics Stock Center (http://www.fgsc.net).
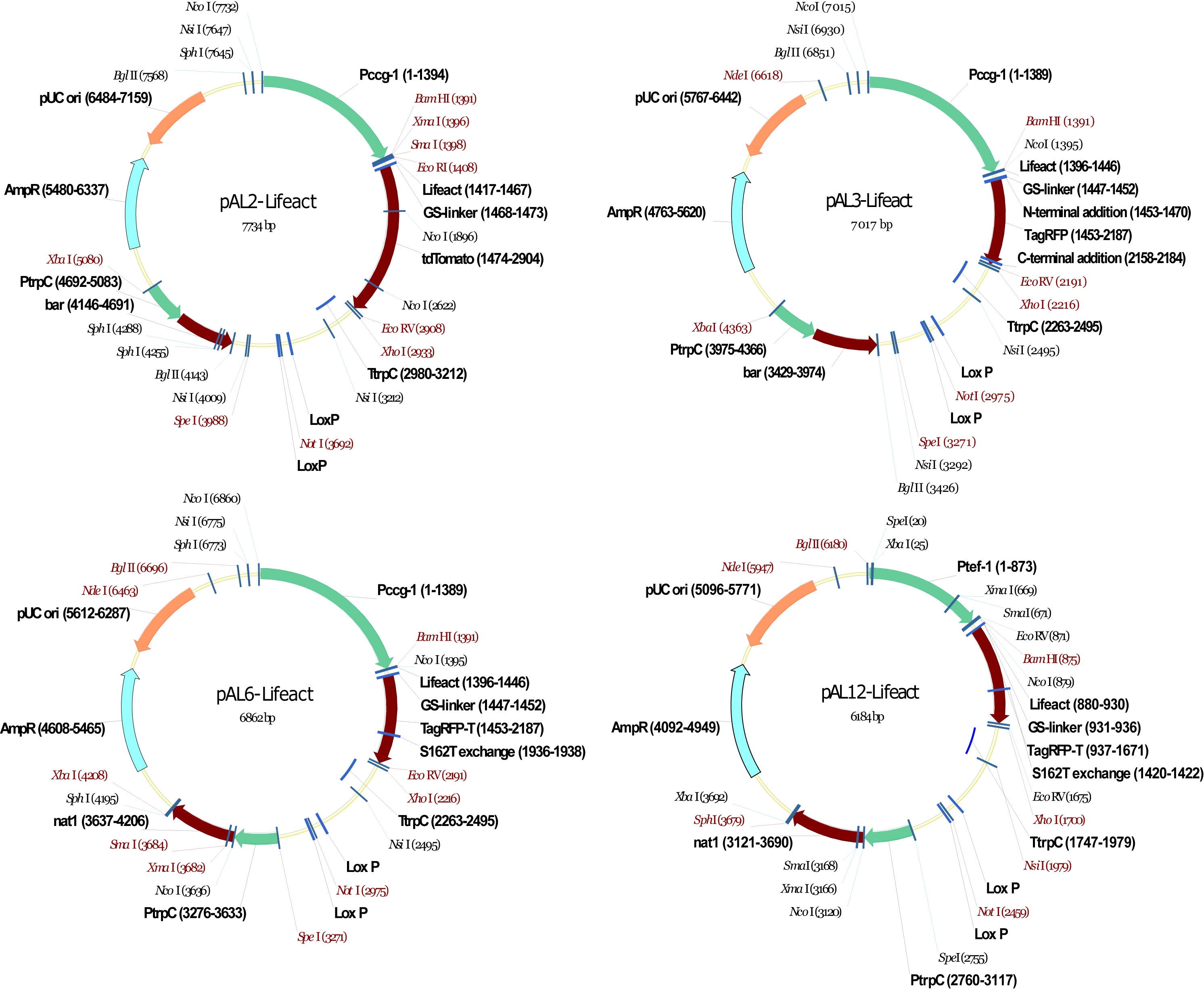
Figure 3. Physical maps of a selection of pALx-Lifeact plasmids. Sequences and maps of all vectors can be downloaded from the FGSC web site (http://www.fgsc.net). Unique restriction sites are indicated in red.
Acknowledgements
Sincere thanks to the named individuals for sharing unpublished results and providing valuable feedback to the manuscript; especially to Amy Gladfelter and Elias Epp. This research was funded by a Biotechnological and Biological Sciences Research Council grant (grant # BB/E010741/1) to N.D.R.
References
Arpaia, G., Loros, J. J., Dunlap, J. C., Morelli, G., Macino, G., 1995. Light induction of the clock-controlled gene ccg-1 is not transduced through the circadian clock in Neurospora crassa. Mollecular and Geneneral Genetics. 247, 157-63.
Berepiki, A., Lichius, A., Shoji, J.-y., Tilsner, J., Read, N. D., 2010. F-actin dynamics in Neurospora crassa. Eukaryotic Cell. 9, 547-57.
Böhmer, C., Ripp, C., Bölker, M., 2009. The germinal centre kinase Don3 triggers the dynamic rearrangement of higher-order septin structures during cytokinesis in Ustilago maydis. Miolecular Microbiology. 74, 1484-1496.
Coffman, V. C., Nile, A. H., Lee, I. J., Liu, H., Wu, J.-Q., 2009. Roles of formin nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Molecular Biology of the Cell. 20, 5195-5210.
Colot, H. V., Park, G., Turner, G. E., Ringelberg, C., Crew, C. M., Litvinkova, L., Weiss, R. L., Borkovich, K. A., Dunlap, J. C., 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proceedings of the National Academy of Sciences of the USA. 103, 10352-10357.
Delgado-Álvarez, D. L., Callejas-Negrete, O. A., Gómez, N., Freitag, M., Roberson, R. W., Smith, L. G., Mouriño-Pérez, R. R., 2010. Visualization of F-actin localization and dynamics with live cell markers in Neurospora crassa. Fungal Genetics and Biology. 47, 573-586.
Era, A., Tominaga, M., Ebine, K., Awai, C., Saito, C., Ishizaki, K., Yamato, K. T., Kohchi, T., Nakano, A., Ueda, T., 2009. Application of Lifeact reveals F-actin dynamics in Arabidopsis thaliana and the liverwort, Marchantia polymorpha. Plant and Cell Physiology. 50, 1041-1048.
Estecha, A., Sanchez-Martin, L., Puig-Kroger, A., Bartolome, R. A., Teixido, J., Samaniego, R., Sanchez-Mateos, P., 2009. Moesin orchestrates cortical polarity of melanoma tumour cells to initiate 3D invasion. Journal of Cell Science. 122, 3492-501.
Freitag, M., Hickey, P. C., Raju, N. B., Selker, E. U., Read, N. D., 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genetics and Biology. 41, 897-910.
Goldstein, A. L., McCusker, J. H., 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 15, 1541-53.
Hays, S., Selker, E. U., 2000. Making the selectable marker bar tighter and more economical. Fungal Genetics Newsletter. 47, 107.
Hentges, P., Van Driessche, B., Tafforeau, L., Vandenhaute, J., Carr, A. M., 2005. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast. 22, 1013-9.
Kawaguchi, Y., Honda, H., Taniguchi-Morimura, J., Iwasaki, S., 1989. The codon CUG is read as serine in an asporogenic yeast Candida cylindracea. Nature. 341, 164-166.
Kinoshita, M., Field, C. M., Coughlin, M. L., Straight, A. F., Mitchison, T. J., 2002. Self- and actin-templated assembly of mammalian septins. Developmental Cell. 3, 791-802.
Kück, U., Hoff, B., 2006. Application of the nourseothricin acetyltrasnferase gene (nat1) as dominant marker for transformation of filamentous fungi. Fungal Genetics Newsletter. 53, 9-11.
Loros, J. J., Denome, S. A., Dunlap, J. C., 1989. Molecular cloning of genes under control of the circadian clock in Neurospora. Science. 243, 385-388.
Maerz, S., Dettmann, A., Ziv, C., Liu, Y., Valerius, O., Yarden, O., Seiler, S., 2009. Two NDR kinase-MOB complexes function as distinct modules during septum formation and tip extension in Neurospora crassa. Molecular Microbiology. 74, 707-723.
Maerz, S., Ziv, C., Vogt, N., Helmstaedt, K., Cohen, N., Gorovits, R., Yarden, O., Seiler, S., 2008. The nuclear Dbf2-related kinase COT1 and the mitogen-activated protein kinases MAK1 and MAK2 genetically interact to regulate filamentous growth, hyphal fusion and sexual development in Neurospora crassa. Genetics. 179, 1313-1325.
McDade, H. C., Cox, G. M., 2001. A new dominant selectable marker for use in Cryptococcus neoformans. Medical Mycology. 39, 151-4.
McNally, M. T., Free, S. J., 1988. Isolation and characterization of a Neurospora glucose-repressible gene. Current Genetics. 14, 545-551.
Metzenberger, R. L., Jacobson, D. J., Bertrand, H., 2000. Making the selective agent for the Bar plasmids, phosphinothricin (glufosinate) affordable for routine use. Fungal Genetics Newsletter. 47, 79-80.
Pall, M. L., Brunelli, J. P., 1994. New plasmid and plasmid hybrid vectors and a Neurospora crassa genomic library containing the bar selectable marker and Cre/lox site-specific recombination system for use in filamentous fungi. Fungal Genetics Newsletter. 41, 63-65.
Read, N. D., Lichius, A., Shoji, J., Goryachev, A. B., 2009. Self-signalling and self-fusion in filamentous fungi. Current Opinion in Microbiology. 12, 608-615.
Riedl, J., Crevenna, A. H., Kessenbrock, K., Yu, J. H., Neukirchen, D., Bista, M., Bradke, F., Jenne, D., Holak, T. A., Werb, Z., Sixt, M., Wedlich-Söldner, R., 2008. Lifeact: a versatile marker to visualize F-actin. Nature Methods. 5, 605-607.
Riedl, J., Flynn, K. C., Raducanu, A., Gartner, F., Beck, G., Bosl, M., Bradke, F., Massberg, S., Aszodi, A., Sixt, M., Wedlich-Soldner, R., 2010. Lifeact mice for studying F-actin dynamics. Nature Methods. 7, 168-9.
Roca, M. G., Kuo, H.-C., Lichius, A., Freitag, M., Read, N. D., 2010. Nuclear dynamics, mitosis and the cytoskeleton during the early stages of colony initiation in Neurospora crassa. Eukaryotic Cell. 9, 1171-1183.
Shaner, N. C., Campbell, R. E., Steinbach, P. A., Giepmans, B. N. G., Palmer, A. E., Tsien, R. Y., 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnology. 22, 1567-1572.
Shaner, N. C., Lin, M. Z., McKeown, M. R., Steinbach, P. A., Hazelwood, K. L., Davidson, M. W., Tsien, R. Y., 2008. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nature Methods. 5, 545-551.
Shen, J., Guo, W., Kohler, J. R., 2005. CaNAT1, a heterologous dominant selectable marker for transformation of Candida albicans and other pathogenic Candida species. Infection and Immunity. 73, 1239-1242.
Smertenko, A. P., Deeks, M. J., Hussey, P. J., 2010. Strategies of actin reorganisation in plant cells. Journal of Cell Science. 123, 3019-28.
Vidali, L., Rounds, C. M., Hepler, P. K., Bezanilla, M., 2009. Lifeact-mEGFP reveals a dynamic apical F-actin network in tip growing plant cells. PLoS ONE. 4, e5744.