Analysis of the DNA
sequence of the putative ABC transporter NCU09975 in Neurospora crassa strains carrying acriflavin resistance markers.
Aric
E. Wiest, Sarah Koch and Kevin McCluskey. Fungal Genetics Stock Center,
University of Missouri- Kansas City, Kansas City, MO 94110 USA
Fungal
Genet Reports 59: 26 - 29
Genomic DNA sequence was determined for the putative Neurospora crassa ABC transporter NCU09975 from several different
classical mutant strains including several acriflavin resistant mutants. The sensitivity of these strains to
acriflavin was tested. While the open reading frame NCU09975 has multiple
polymorphisms in strains sequenced for other purposes, none of the acriflavin
resistant classical mutants tested had polymorphisms in the NCU09975 coding
region or in the 195 bases upstream of the translation start site.
Introduction
While
years of mutagenesis and subsequent characterization in Neurospora crassa
produced one of the most densely saturated genetic maps (Perkins et al. 2001),
many of the classical mutants are not yet associated with open reading frames
from the genome sequence (Galagan et al. 2003). Among these are acriflavin
resistant mutants which have been assigned to seven different loci. A subset of
these genes also confer resistance to multiple related drugs, including acridine orange, malachite green, and aminotriazole
(Akiyama and Nakashima 1996). Moreover
some acriflavin resistance genes are dominant while others are recessive. Because we wanted to find a dominant
selectable marker for transformation of Neurospora and because the abc-3-like
ORF NCU09975 had a large number of polymorphisms in its primary sequence among
a group strains subject to whole genome sequence (McCluskey et al. 2011) we
undertook to characterize this locus in strains carrying the acriflavin
resistance markers acr-1 and Acr-3. We additionally validated the acriflavin
sensitivity in the classical mutant strains carrying polymorphisms in NCU09975.
Materials and
methods
Strains
were cultured on Vogel's minimal medium (Vogel 1956) using standard practices
(Davis and De Serres 1970) (Table 1).
Table 1. Strains and their
characteristics
|
FGSC
number |
Genotype |
Marker
Location |
Acriflavin
sensitivity |
|
2489 |
mat-A |
IL |
Sensitive |
|
1215 |
Acr-3 mat-a |
IL IL |
Resistant |
|
1209 |
Acr-3
mat-A |
IL IL |
Resistant |
|
875 |
acr-1 mat-a |
IL IL |
Sensitive |
|
305 |
amyc
mat-A |
IL IL |
Sensitive |
|
1363 |
smco-1
mat-A |
I IL |
Sensitive |
|
3921 |
tng
mat-A |
IIL; IL |
Sensitive |
|
7022 |
fld |
IVR; IL |
Sensitive |
Acriflavin
was dissolved in water and filter sterilized prior to addition to sterile
culture medium. It was stored in the dark and fresh stocks were prepared
regularly. Acriflavin sensitivity
testing was carried out by pipetting 10
ul of a freshly prepared suspension of conidia and hyphal fragments in water
(approximately 103cfu/ ml) onto the surface of agar solidified
medium in 10 x 75 mm glass culture tubes. Results were logged after two, four
and ten days. Genomic DNA was extracted
from 2 - 3 day old liquid shake cultures using the ZR Fungal DNA kit (Zymo Research, Irvine, CA).
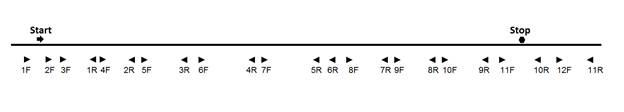
NCU09975 DNA was amplified as shown in Figure 1, using primers specific
for this ORF (Table 2). Primers 1F and 5R were used together to amplify a 2848
base fragment including the start codon and 195 upstream bases. Primers 7F and
11R were used to amplify a 2878 base fragment including the stop codon and 667
bases downstream (Figure 1). These large fragments which overlap by 370 bases
were directly sequenced at the UMKC School of Biological Sciences core facility
using an Applied Biosystems 3100 Genetic Analyzer
(Foster City, USA). Sequences were aligned and analyzed using Sequencher (Gene Codes Corporation, Ann Arbor, USA).
Table 2. Primers
|
Primer Designation and
Orientation |
Position |
Sequence |
|
1F |
-195 |
ATTCGTCTCGACTTGCGACT |
|
2F |
24 |
TTCGTTGTCACTCGTCTTGG |
|
3F |
306 |
CGTTTGAGTTGGCGATCA |
|
4F |
784 |
AGCAGCCCTGACTTGCAT |
|
5F |
1303 |
CCTTCTTCGCTGCCTTTG |
|
6F |
1813 |
TCATCGACCGCAAGTCAA |
|
7F |
2283 |
AGCGGCTCGTTGAAGATG |
|
8F |
2784 |
AAATTGGAGGAGCTGCGATA |
|
9F |
3277 |
TCCGCTTCTATCGGTACACA |
|
10F |
3778 |
TCGATGGCATTGGGTTTT |
|
11F |
4295 |
CCACCGGGTCAGTTTGTC |
|
12F |
4786 |
CGAAGGTGGAGCAGATGG |
|
1R |
641 |
TGAGGGGTCTCGTCTTCCT |
|
2R |
1135 |
ATCCTGATGGCGTTGGTG |
|
3R |
1634 |
TCCAGCGTACACGCAAAA |
|
4R |
2149 |
CCTGCATGCTTACCTGCTG |
|
5R |
2653 |
CGACCGTTGGACATGACA |
|
6R |
2750 |
CTTTTGGGGTTCGTCATCAT |
|
7R |
3163 |
AGTTGCCAAGGGCTACGA |
|
8R |
3635 |
TGGCCATGATAGCCTCAGA |
|
9R |
4136 |
CGCCGTTGCTACGTTTTT |
|
10R |
4628 |
GAGATCCGCAGGGGGTAG |
|
11R |
5161 |
GACGGAGATGACCCGAAA |
Results and
Discussion
The primary sequence of NCU09975 was identical to wild type in strains FGSC 1215 and FGSC 1209 (Acr-3) and also in strain FGSC 875 (acr-1, Table 1). Strain FGSC 1215 and FGSC 1209 were both resistant to acriflavin at the highest concentration tested (50 ug/ml). Strain FGSC 875 which caries the acr-1 mutation was sensitive to acriflavin at all concentrations above 2.5 ug/ ml, as was the reference genome strain FGSC 2489 (74-OR23-1VA, Table 1 and Figure 2).
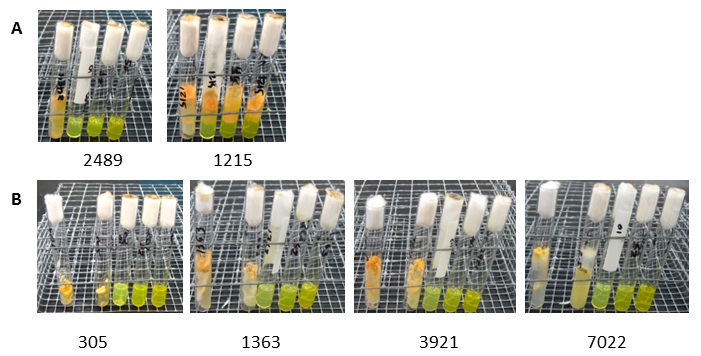
Figure 2. Acriflavin sensitivity tests A)
Wild type (2489) and Acr-3 mutant
strains (1215) of Neurospora crassa.
Left to right (in each test tube): 2.5 ug/ml acriflavin, 10 ug/ml, 25 ug/ml, 50
ug/ml. B) Acriflavin sensitivity of classical mutant strains of N. crassa. Left to right (in each test
tube): 0 ug/ml acriflavin, 2.5 ug/ml, 10 ug/ml, 25 ug/ml, 50 ug/ml.
Single
Nucleotide Polymorphisms (SNPs) in NCU09975 were detected in four strains
subject to whole genome sequence as part of another study (McCluskey et al.
2011). These strains, FGSC 305, 1363, 3921 and 7022, all have mutations
unrelated to their potential response to acriflavin (Table 1). While most of
the polymorphisms at NCU09975 in these strains are shared among all four
strains, strain 3921 has ten unique SNPs and strain 7022 has two unique
SNPs. Both of the unique SNPs in FGSC
7022 are in the ninth intron. The unique SNPs in strain 3921 include four
synonymous SNPs, four non-synonymous SNPs and one SNP each in intron one and
two, respectively. Among the shared SNPs in these strains are fourteen that are
shared among multiple strains. One SNP is present in two strains, ten SNPs are
found in three strains and three SNPs are found in all four strains. Eight of
these SNPs are synonymous while three are non-synonymous and three are found in
introns. None of these strains had any insertions or deletions in NCU09975. All
four of these strains were sensitive to acriflavin at 10 ug/ ml, as was the
wild type strain FGSC 2489 (Figure 2). Strain FGSC 305 contains the
morphological mutation amyc
which causes it to grow as a small
dot-like colony (Perkins 1959) and while this makes comparison to wild-type
difficult, the comparison between growth of this strain on Vogels
medium without acriflavin and with acriflavin was straightforward (Figure 2B).
Strains FGSC 1363, 3921 and 7022 each
carry morphological mutations (Table 1), but these did not impact the ability
to score their growth on acriflavin containing medium.
The
acriflavin resistance gene Acr-2
(NCU05733) was previously characterized as a Zn(II)Cys6 binuclear domain
containing protein and disruption of this gene rendered progeny acriflavin
sensitive (Akiyama and Nakashima 1996).
Like Acr-3, acr-4
is also on linkage group IL and it confers resistance to 50 ug/ml acriflavin.
Genetic mapping places acr-4 5 map
units away from Acr-3 (Hsu 1965) and
while Acr-3 is a dominant mutation, acr-4 is recessive. acr-5 is on linkage group II L and unlike other acriflavin
resistance traits does not confer resistance to other drugs such as malachite
green or acridine orange (Akiyama and Nakashima
1996). Some alleles of acr-5 are only
manifest in strains also carrying the morphological marker mo(KH161). acr-6,
on linkage group III R, also confers resistance to acridine
orange, but not to malachite green (Akiyama and Nakashima 1996).
While
this work did not identify the ORF responsible for acriflavin resistance
conferred by mutations at the Acr-3
locus in N. crassa, analysis of
additional ORFs in the region near NCU09975 is ongoing.
Acknowledgements
The FGSC is
supported by grant 742713 from the US National Science Foundation. S. Koch is a
student at the Pembroke Hill School, Kansas City, MO.
References
AKIYAMA,
M., and H. NAKASHIMA, 1996 Molecular cloning of the acr-2 gene which controls
acriflavin sensitivity in Neurospora crassa. Biochim Biophys Acta 1307: 187-192.
DAVIS,
R. H., and F. J. DE SERRES, 1970 Genetic and microbiological research
techniques for Neurospora crassa. Meth. Enzymol. 17:
79-143.
GALAGAN,
J. E., S. E. CALVO, K. A. BORKOVICH, E. U. SELKER, N. D. READ et al., 2003 The
genome sequence of the filamentous fungus Neurospora crassa. Nature 422:
859-868.
HSU,
K. S., 1965 Acriflavin resistance controlled by chromosomal genes in
Neurospora. Neurospora Newsl. 8: 4-6.
MCCLUSKEY,
K., A. WIEST, I. V. GRIGORIEV, A. LIPZEN, J. MARTIN et al., 2011 Rediscovery by
whole genome sequencing: classical mutations and genome polymorphisms in
Neurospora crassa. G3 Genes, genomes, genetics 1: 303-316
PERKINS,
D. D., 1959 New Markers and Multiple Point Linkage Data in Neurospora. Genetics
44: 1185-1208.
PERKINS,
D. D., A. RADFORD and M. S. SACHS, 2001 The Neurospora
compendium : chromosomal loci. Academic press, San Diego, CA.
VOGEL,
H. J., 1956 A convenient growth medium for Neurospora (Medium N). Microbial.
Genet. Bull. 13: 42-43.