Efficient tools to target DNA to
Podospora anserina
Michelle Déquard-Chablat1,2, Tan-Trung Nguyen1,2,
Véronique Contamine1,2, Sylvie Hermann-Le Denmat1,2,3 and
Fabienne Malagnac1,4.
1
Univ Paris-Sud, Institut de Génétique et Microbiologie, UMR 8621, ORSAY,
F-91405, France. 2 CNRS, ORSAY, F-91405, France.
3
Ecole Normale Supérieure, 75005 Paris, France.
4
Univ Paris Diderot, Sorbonne Paris Cité, UFR des Sciences du Vivant, 75205 Paris
CEDEX 13, France
(pdf)
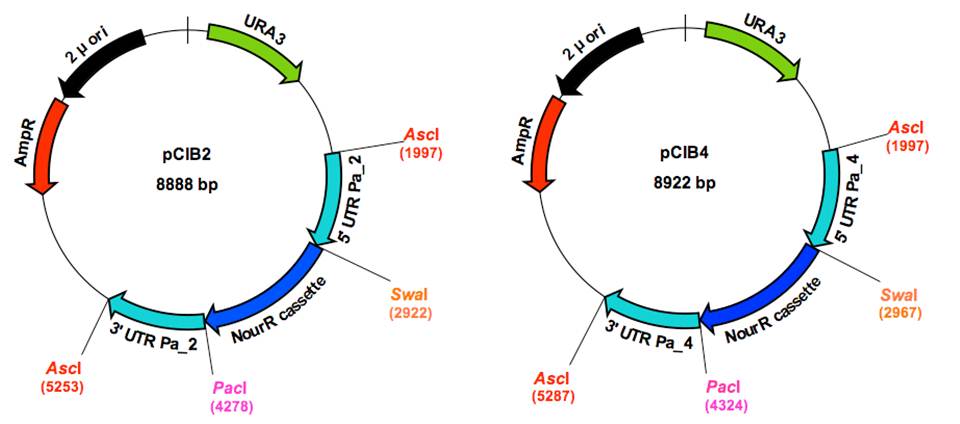
pCIB2 and pCIB4 plasmid constructions
As for Neurospora crassa, we set up a yeast recombinational cloning
strategy using the pRS426 shuttle plasmid (Colot et al., 2006). For selectable
marker, we chose the nourseothricine resistance gene (NouR) from pAPI508 (El-Khoury
et al., 2008). First, we PCR-amplified from plasmid genomic libraries (Espagne
et al., 2008) the flanking 5-prime and 3-prime regions of Pa_2_3690
(VeA5f2/NoS5r2 and NoP3f2/VeA3r2) and Pa_4_5450 (VeA5f4/NoS5r4 and
NoP3f4/VeA3r4) respectively (about 1 kb each). Primers NoS5r2 and NoS5r4 contain
a SwaI restriction site, whereas primers NoP3f2 and NoP3f4 contain a PacI
restriction site. These two enzymes were chosen for restriction cloning because
they recognize eight-base sequences, which are very infrequent in the Podospora
genome. The flanking regions do not encompass coding sequence of the neighboring
genes. Primers were designed to overlap either with pRS426 (VeA5f2, VeA3r2 and
VeA5f4, VeA3r4) or with the NouR cassette (NoS5r2, NoP3f2 and NoS5r4, NoP3f4).
Meanwhile, the NouR cassette was amplified using primers No5f, No3r. All PCR
were done using the high fidelity Pfu DNA polymerase (Promega) for 25 cycles of
amplification.
A W303 yeast strain was transformed with the 5-prime and 3-prime flanking
regions as well as the NouR cassette and the pRS426 plasmid digested with XhoI
and EcoRI for each construction. Because of homologous ends, the four fragments
were ligated, generating the pCIB2 (Pa_2_3690) and pCIB4 (Pa_4_5450) plasmids.
For each construction, four yeast colonies showing restored prototrophy to
Uracil were selected. Plasmid DNA was extracted and used as template for PCR
amplification (primers PU/NouR Int Rev and RP/NouR Int) to check the
constructions. Once verified, the plasmids were transformed into Escherichia
coli. Ten clones were picked for each transformation and checked by colony
PCR with the primers mentioned above. pCIB2 (Genbank accession number JX262692)
and pCIB4 (Genbank accession number JX262693) plasmid DNA was purified from two
large cultures, inoculated with a single clone and sequenced (see Tables 1 and 2
for primers and Figure 1 for plasmid graphic maps). pCIB2 and pCIB4 are
available from the Fungal Genetics Stock Center (Kansas City, MO).
Table1:
Primers used to amplify the flanking regions of the target genes and the NouR
cassette
|
name |
5-prime -> 3-prime sequence |
|
No5f
|
tcttcccttccacttcttcacacagaccac |
|
No3r
|
ggcacaaagcatcaagaaggcaaacagaac |
|
Pa_2_3690 locus |
|
|
VeA5f2
|
gagcgcgcgtaatacgactcactatagggcgcgccaccggtgttctcggcatctttctg |
|
NoS5r2
|
gtggtctgtgtgaagaagtggaagggaagatttaaatcgtctgacccgatcaaagcatgag |
|
NoP3f2
|
gttctgtttgccttcttgatgctttgtgccttaattaaccgcatgtttggtgttggc |
|
VeA3r2
|
gaacaaaagctggagctccaccgcggtggcgCgccgcagacagggaaccaaggac |
|
Pa_4_5450 locus |
|
|
VeA5f4
|
gagcgcgcgtaatacgactcactatagggcgcgCcgtgtgcgctaaacgttgggtaatg |
|
NoS5r4
|
gtggtctgtgtgaagaagtggaagggaagatttaaatggtaccttatctggcctgtcc |
|
NoP3f4 |
gttctgtttgccttcttgatgctttgtgccttaattAAggtcacaccgaactgagaagg |
|
VeA3r4
|
gaacaaaagctggagctccaccgcggtggcgCgccacaccttcgcagacctagc |
Sequences in bold, regular and underlined correspond respectively to the pRS426
vector, the ends of the resistance cassette and the ends of the flanking region
of one of each target gene. Bases in italic uppercase were added to create
appropriate restriction sites (ggcgcgcc =
AscI, atttaaat = SwaI, ttaattaa =
PacI).
Table 2:
Primers used to check the constructions and the integration in Podospora genomic
DNA.
|
name |
5-prime -> 3-prime sequence |
position |
|
Ext 5’2 |
gttctgcctcaccttcatcg |
Upstream of Pa_2_3690 5’UTR |
|
Ext 3’2 rev |
acgtggccctgacatcatc |
Downstream of Pa_2_3690 3’UTR |
|
|
|
|
|
Ext 5’4 |
tccacagcctacgacaggtg |
Upstream of Pa_4_5450 5’UTR |
|
Ext 3’4 rev |
cccttcggtgaactacctg |
Downstream of Pa_4_5450 3’UTR |
|
NouR Int |
ttcgtggtcgtctcgtactc |
Inside
Nat1 gene |
|
NouR Int Rev |
ggtgcgttgacgttggtgac |
Inside
Nat1 gene |
Cloning DNA fragments into pCIB2 and pCIB4
To clone alleles of interest into either pCIB2 or pCIB4, blunt-ended DNA
fragments can be ligated to the SwaI site, whereas DNA fragments with cohesive
ends generated by PvuI or AsiSI digestions can be ligated to PacI site. Prior to
transformation, the ligations were hydrolyzed using SwaI or PacI, in order to
eliminate self-ligated vectors.
Once alleles of interest are introduced into pCIB2 and pCIB4, and prior to
transformation into Podospora, these plasmids have to be digested to generate
linear fragments to favor homologous recombination at the corresponding target
locus. For this purpose AscI sites have been introduced into primers VeA5f2,
VeA3r2 and VeA5f4, VeA3r4 (see Figure 1).

As a proof of principle, site-directed integration was tested. Alleles of four
different genes were cloned into pCIB4, using either SwaI or PacI. After
independent transformations of the ΔPaKu70 strain (El-Khoury et al., 2008) with
five linear fragments, PCR analyses of a total of 30 transformants showed 100%
of single-copy integration at the Pa_4_5450 targeted locus. We then checked the
expression of the five transgenes. We first saw that introduction of wild-type
allele of Pa_3_6770 restores viability of the corresponding deleted mutant,
which proves efficient transcriptional gene expression at the Pa_4_5450 locus.
When Pa_3_6770 was integrated at the same Pa_4_5450 locus but driven by the
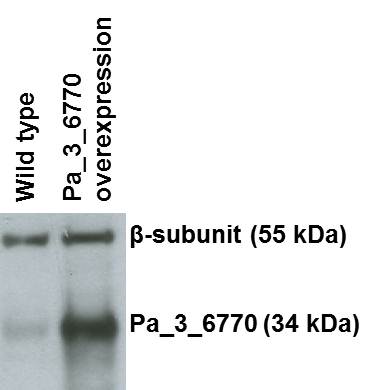
promoter of the highly expressed AS4 gene (Silar & Picard, 1994), it showed a
14-fold increase in expression (as measured by RT-qPCR). Western-blot analysis
further confirmed this over-expression (Figure 2). Additional RT-qPCR
experiments performed on the remaining three over-expressed alleles integrated
at the Pa_4_5450 locus also showed clear increases, ranging from 6- to 50-fold.
The Podospora community can now rely on an efficient targeting system to compare
the expression of various alleles of a given gene.
Acknowledgments
We thank Robert Debuchy, Evelyne Coppin and Jinane Aït Benkhali for discussion
and Robert Debuchy and Vinosa Yogarajah for technical assistance.
We also thank Jean Velours (IBGC-CNRS, Bordeaux, France) and Geneviève Dujardin
(CGM-CNRS, Gif/Yvette, France) for providing the
S. cerevisiae anti-β-subunit antibody
and Pasquale Scarcia and Ferdinando Palmieri (University of Bari, Italy) for
providing antibodies directed against Pa_3_6770
S. cerevisiae homologous polypeptide.
References
Bidard F, Aït Benkhali J, Coppin E, Imbeaud S, Grognet P, Delacroix H and
Debuchy R, 2011. Genome-wide gene expression profiling of fertilization
competent mycelium in opposite mating types in the heterothallic fungus
Podospora anserina. PLoS ONE
6: e21476.
Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL,
Borkovich KA and Dunlap JC, 2006. A high-throughput gene knockout procedure for
Neurospora reveals functions for multiple transcription factors. Proc. Natl.
Acad. Sci. U.S.A. 103:
10352-10357.
El-Khoury R, Sellem CH, Coppin E, Boivin A, Maas MFPM, Debuchy R and
Sainsard-Chanet A, 2008. Gene deletion and allelic replacement in the
filamentous fungus Podospora anserina.
Curr. Genet.
53: 249-258.
Espagne E, Lespinet O, Malagnac F, Da Silva C, Jaillon O, Porcel BM, Couloux A,
Aury J-M, Ségurens B, Poulain J, et al.,
2008.
The genome sequence of the model ascomycete fungus
Podospora anserina. Genome Biol
9: R77.
Silar P and Picard M, 1994. Increased longevity of EF-1 alpha high-fidelity
mutants in Podospora anserina.
J. Mol. Biol.
235: 231-236.