Differential
sensitivity of Ustilago maydis to fungal
antibiotics on simple and complex media
Anju Verma1, Tamas Kapros2, and Jakob
H. Waterborg2 *
1 Division of Plant Sciences and
Bond Life Sciences Center, University of Missouri-Columbia, Columbia, MO 65211,
USA; 2 Division of Cell
Biology and Biophysics, School of Biological Sciences, University of
Missouri-Kansas City, Kansas City, MO 64110, USA.
* Corresponding author
Fungal Genetics Reports 62:8- 13
(pdf)
We have observed that the basidiomycete Ustilago maydis can be partially or completely resistant to
antibiotics when grown in defined growth media.
In synthetic medium based on the fully defined mixture of simple organic
compounds and salts U. maydis
displays near wild-type growth at concentrations of hygromycin that effectively
kill cells in complex nutrient media. The
antibiotics geneticin, nourseothricin and phleomycin had similar effects. In contrast, the fungicide carboxin was
equally effective in all growth media tested.
Our observations could guide selection of growth media for genetic
transformation of Ustilago and other
fungi when sensitivity to common antibiotics is used as a selectable marker.
Introduction
Like many other laboratories we have successfully used the
fungicide carboxin as a selectable marker in gene transformation (Brachmann et al., 2004; Brachmann et al., 2001;
Fernandez-Alvarez et al., 2009; Kojic and Holloman, 2000; Topp et al., 2002)
to knock-out variant-specifically histone H3 isotype loci (Verma et al., 2011). After transformation, transformants were
cultured under continued selection until stable, non-heterokaryon clones could
be isolated (Verma et al., 2011). These cultures were maintained on synthetic
minimal media consisting of yeast nitrogen base (YNB), a standard mixture of small
organic compounds and salts, with glucose (SD).
When attempting to use other standard antibiotic selectable markers such
as hygromycin, geneticin, nourseothricin and phleomycin (Brachmann et al., 2004; Kamper, 2004; Kojic and Holloman, 2000),
selection was not observed. Other
laboratories have used these markers extensively in generating genetic
transformants of Ustilago, but they
used chemically complex media, primarily based on mixtures of yeast extract and
bactopeptone supplemented with glucose (YPD) or sucrose (YPS) (Berndt et al., 2010; Brachmann et al., 2004;
Brachmann et al., 2001; Lovely et al., 2011; Tsukuda et al., 1988), but
also Complete Medium (Garcia-Pedrajas et al.,
2008; Holliday, 1974; Lee et al., 1999) or Potato Dextrose (Brachmann et al., 2004; Heidenreich et al., 2008;
Zameitat et al., 2007). In our hands
too, these complex media, in particular YPS, can be used effectively for
antibiotic selection. However, we report
here that in simple SD medium aminoglycoside and glycopeptide antibiotics, all
larger and more hydrophilic than carboxin, are largely or completely
ineffective. We speculate that uptake of
these antibiotics depends on Ustilago
cell membrane transport mechanisms that are absent in the simple SD medium. This observation poses an impediment if one
wants to create auxotrophic mutants, e.g. for essential amino acids like
leucine (Fotheringham and Holloman, 1990)
or nucleotide bases like adenine (Verma et al.,
2016) in YNB-based synthetic minimal media.
Materials
and Methods
Simple, synthetic-defined media SD and SS consisted of 6.7 g
yeast nitrogen base (YNB) without amino acids (Fisher Scientific, BD Difco) with
20 g dextrose (glucose) or sucrose (Sigma-Aldrich), respectively, in 1 L
water. pH-buffered SD medium (SDS) was
prepared by including 50 mM succinic acid and adjustment of the pH to 7 with
NaOH. Semi-defined, complex growth media
YPD and YPS2 contained 10 g yeast extract (BD Difco), 20 g bactopeptone (BD Difco)
with 20 g glucose or sucrose, respectively, in 1 L water. In addition to this rich complex medium (Tsukuda et al., 1988), we also tested the
poorer formulation YPS1 with only 4 g yeast extract and 4 g bactopeptone per L (Brachmann et al., 2004). Ustilago
growth rates and antibiotic effects were indistinguishable between YPS1 and
YPS2. For plate cultures, media were
supplemented with 15 g agar (Fisher Scientific) per L. Unless specified, media were autoclaved without
adjusting the pH of approximately 6.5.
Aliquots of stock solutions of antibiotics were added to the
desired final concentrations into autoclaved simple or complex agar media, after
cooling to approximately 55 C.
Filter-sterilized hygromycin (Sigma-Aldrich) and geneticin (G418, Gibco
BRL) stocks were 50 mg/mL water; nourseothricin (NTC, Jena Bioscience, Germany)
was 100 mg/mL water; phleomycin (InVivogen) was 20 mg/mL stock as purchased. Carboxin (5,6-dihydro-2-methyl-1,4-oxathi-ine-3-carboxanilide;
Vitavax) was 10 mg/mL methanol, diluted from a 34% suspension in methanol, a
kind gift from S. Gold (Athens, GA) and used at 3 μg/mL (Verma
et al., 2011).
Wildtype U. maydis
521 haploid strain FGSC 9021 (Verma et al, 2011) was revived from –70 C
storage in 50% glycerol, grown on SD plates at 30 C and single colonies
were grown overnight in 50 mL SD or YPD at 30 C at 150 rpm in 125 mL
flasks. Multiple samples of 250 cells in
10 microliter medium, after volume adjustment based on hematocytometer
counting, were spotted on 100 mm diameter Petri dishes with 40 ml agar medium,
allowed to dry, and incubated in the dark at 30 C for at least 1 week. Note that initial inoculum sizes increased as
colonies grew, spreading across the agar surface (Fig. 1).
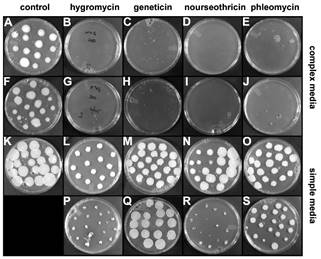
Figure 1. Examples of U. maydis growth on solid media. Multiple aliquots of 10 µL of Ustilago suspension culture containing
250 cells were spotted on agar plates with complex or simple nutrient media and
with sucrose or glucose (see Legend of Table 1) and grown in the dark at
30 C for 3 (A, F) or 6 days (B-E, G-S).
A-J: complex media; K-S: simple media.
Table 1 scores are indicated between square brackets. A.
control, YPS1 [+++]; B. hygromycin,
200 μg/mL,
YPS1 [NO]; C. geneticin, 200 μg/mL,
YPS1 [NO]; D. nourseothricin, 150 μg/mL,
YPS1 [NO]; E. phleomycin, 50 μg/mL,
YPS1 [NO]; F. control, YPS2 [+++]; G. hygromycin, 300 μg/mL,
YPS1 [NO]; H. geneticin, 250 μg/mL,
YPS2 [NO]; I. nourseothricin, 300 μg/mL,
YPS2 [NO]; J. phleomycin, 20 μg/mL,
YPS2 [NO]; K. control, SD [+++]; L. hygromycin, 150 μg/mL,
SD [++]; M. geneticin, 100 μg/mL,
SD [+++]; N. nourseothricin, 150 μg/mL,
SD [++]; O. phleomycin, 15 μg/mL,
SD [+++]; P. hygromycin, 250 μg/mL,
SD [+]; Q. geneticin, 300 μg/mL,
SD [+++]; R. nourseothricin, 300 μg/mL,
SD [+]; S. phleomycin, 50 μg/mL,
SD [++]. Note the somewhat variable size
of the 10 μL
spots applied on plates like A where large colonies grow in only 3 days and
plates like B, where the application spots remain faintly visible but
microscopic colonies are never detected.
Results
and Discussion
In our research to create selective knock-outs for the two
single locus histone H3 variants in Ustilago,
successful transformation at single loci was achieved using the selectable
marker carboxin on simple defined (SD) growth media based on yeast nitrogen
base (YNB) and glucose (Verma et al., 2011). The mutant succinate dehydrogenase cassette (Brachmann et al., 2004) which confers
resistance to carboxin (Keon et al., 1991)
was stably integrated in the Ustilago
genome at the knock-out locus. To study
the single remaining histone H3 variant function, we wished to replace its
promoter by that of the inactivated gene, switching between the cell-cycle regulated
H3 promoter and the constitutive H3 promoter (Verma
et al., 2011).
In the search for a second usable selectable marker, we were
surprised by abundant growth on SD plates with hygromycin, geneticin (G418),
nourseothricin (NTC) and phleomycin (Fig. 1) at antibiotic concentrations that
were described as effective by other laboratories using Ustilago (Berndt et al., 2010;
Brachmann et al., 2004; Brachmann et al., 2001; Garcia-Pedrajas et al., 2008;
Heidenreich et al., 2008; Kamper, 2004; Kojic and Holloman, 2000; Lee et al.,
1999; Lovely et al., 2011; Tsukuda et al., 1988; Zameitat et al., 2007). Reviewing growth media used by these
laboratories, none had used simple media like our YNB-based one with
glucose. We confirmed that indeed these
antibiotics were effective against wild type Ustilago strains when yeast extract- and bactopeptone-based media
were used (Fig. 1) at the effective concentrations described (Table 1).
We explored some of the possible factors involved to find
conditions that would allow use of these antibiotics as selectable markers in
simple media during the development of auxotrophic markers, such as the ade2 knockout (Verma et al., 2016). The
effects of the composition of growth media during genetic transformation and
selection of Ustilago has been
recognized by others. For instance, the
inclusion of high concentrations of sorbitol in protoplast transformation of Ustilago is required for the
stabilization of the protoplasts but it reduces the effectiveness of antibiotic
selection (Kojic and Holloman, 2000).
Table 1. U. maydis growth on solid media.
|
Medium components (g/L) |
|||||||
|
Medium |
yeast extract |
bacto-peptone |
YNB |
sucrose |
glucose |
succinic acid |
|
|
YPS1 |
4 |
4 |
|
20 |
|
|
|
|
YPS2 |
10 |
20 |
|
20 |
|
|
|
|
YPD |
10 |
20 |
|
|
20 |
|
|
|
SS |
|
|
6.7 |
20 |
|
|
|
|
SD |
|
|
6.7 |
|
20 |
|
|
|
SDS |
|
|
6.7 |
|
20 |
5.9 |
|
Table 1, U. maydis growth on solid media, continued.
|
|
Antibiotics (μg/mL) |
||||||||||
|
Medium |
Control |
Hygromycin |
Geneticin |
Nourseothricin |
Phleomycin |
Carboxin |
|||||
|
|
0 |
150 |
200-250 |
300 |
100-300 |
100 |
150 |
250-300 |
10-20 |
30-50 |
3 |
|
YPS1 |
+++ |
|
NO |
NO |
NO |
NO |
NO |
NO |
NO |
NO |
|
|
YPS2 |
+++ |
|
NO |
NO |
NO |
NO |
NO |
NO |
NO |
NO |
NO |
|
YPD |
+++ |
NO |
NO |
|
|
NO |
NO |
|
|
NO |
|
|
SS |
+++ |
++ |
+ |
|
|
+ |
(+) |
|
|
+ |
NO |
|
SD |
+++ |
++ |
+ |
(+) |
+++ |
++ |
++ |
+ |
+++ |
++ |
NO |
|
SDS |
+++ |
+++ |
++ |
|
|
NO |
NO |
|
|
(+) |
|
Complex agar media (YPS1, YPS2, YPD) with yeast extract and
bactopeptone and simple agar media (SS, SD, SDS) with yeast nitrogen base
containing sucrose, glucose and/or succinate buffer (see Materials and
Methods). Growth at 30 C in dark. Examples of scored plates are shown in Fig.
1. Scores: +++ : sizable colonies by day
3 as in untreated controls; ++ : visible colonies by day 3; + : small
macroscopic colonies by day 6 (+) : microscopic colonies by day 6; NO : no
microscopic colonies after 6 or more days.
Empty table fields represent combinations of medium and antibiotic that
were not tested.
The uptake of sugars by fungi is generally mediated by sugar
transporters (Goncalves et al., 2016) and
specific sucrose (MW 342 D) transporters have been identified in Ustilago (Wahl
et al., 2010). That sucrose
transporters may contribute to the entry of the only slightly larger
antibiotics is suggested by lesser growth when antibotics are present in simple
medium with sucrose (SS) than with glucose (SD) (Table 1). The aminoglycosides hygromycin (528 D),
geneticin (497 D) and nourseothricin (multiple components between 499 and 883
D) are only marginally larger than sucrose, although the glycopeptide
phleomycin is significantly larger (1428 D).
Note that all these antibiotics contain amines that would create
increasingly more positively charged antibiotics as cells grow and the
unbuffered media acidify. That the
charged state may impede cell entry is suggested by the observation that
nourseothricin and phleomycin in pH-buffered medium (SDS) are more effective
(Table 1). The smaller molecular weight
(235 D) but especially the hydrophobic character of carboxin is probably
responsible for its effective uptake and action in both simple and complex media
(Table 1).
The major difference observed in this study is the limited
antibiotic effectiveness in the simple media in which the organic components
all have molecular weights less than 450 D, and the complex media with
effective antibiotic action and nutrient molecules in yeast extract and
bactopeptone with molecular weights that are much larger (Fig. 1; Table 1). The requirement of nutrient uptake from these
media will certainly activate cellular import mechanisms, transmembrane
transporters or endocytosis, that will also facilitate the effective uptake of
antibiotics into the cells.
Conclusions
Using aminoglycoside and glycopeptide antibiotics as
selectable markers in gene transformation of fungi like the basidiomycete Ustilago maydis, one must employ growth
media that will induce uptake of complex nutrients to assure concomitant uptake
of antibiotics to confer effective selection.
In minimal media, Ustilago cells
exclude aminoglycoside antibiotics to such a degree that effective selection of
antibiotic-resistant transformants cannot be achieved.
Acknowledgements
This research was supported by the Missouri Life Sciences
Research Board, award 13254 to JHW.
References
Berndt, P., Lanver, D., and Kahmann, R., 2010. The AGC Ser/Thr kinase
Aga1 is essential for appressorium formation and maintenance of the actin
cytoskeleton in the smut fungus Ustilago
maydis. Mol. Microbiol. 78:1484-1499.
Brachmann, A., Konig, J., Julius, C., and Feldbrugge, M., 2004. A
reverse genetic approach for generating gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 272:216-226.
Brachmann, A., Weinzierl, G.,
Kamper, J., and Kahmann, R., 2001. Identification
of genes in the bW/bE regulatory cascade in Ustilago
maydis. Mol. Microbiol. 42:1047-1063.
Fernandez-Alvarez, A., Elias-Villalobos, A., and Ibeas, J.I., 2009. The
O-mannosyltransferase PMT4 is essential for normal appressorium formation and
penetration in Ustilago maydis. Plant
Cell 21:3397-3412.
Fotheringham, S., and Holloman, W.K., 1990. Pathways of transformation
in Ustilago maydis determined by DNA
conformation. Genet. 124:833-843.
Garcia-Pedrajas, M.D., Nadal, M.,
Kapa, L.B., et al., 2008. DelsGate, a robust and rapid gene deletion
construction method. Fungal Genet. Biol. 45:379-388.
Goncalves, C., Coelho, M.A., Salema-Oom, M., and Goncalves, P., 2016.
Stepwise functional evolution in a fungal sugar transporter family. Mol. Biol.
Evol. 33:352-366.
Heidenreich, M.L., Budde, A.D., Zhiqiang, A., and Leong, S.A., 2008.
Disruption of a yeast ADE6 gene homolog in Ustilago
maydis. Fungal Genetics Reports 55:40-43.
Holliday, R., 1974. Ustilago
maydis, in: King, R.C. (Ed.), Handbook of Genetics. Plenum Press, New York.
Kamper, J., 2004. A PCR-based system for highly efficient generation of
gene replacement mutants in Ustilago
maydis. Mol. Genet. Genomics 271:103-110.
Keon, J.P., White, G.A., and Hargreaves, J.A., 1991. Isolation,
characterization and sequence of a gene conferring resistance to the systemic
fungicide carboxin from the maize smut pathogen, Ustilago maydis. Curr. Genet. 19:475-481.
Kojic, M., and Holloman, W.K., 2000. Shuttle vectors for genetic
manipulations in Ustilago maydis. Can. J. Microbiol.
46:333-338.
Lee, N., Bakkeren, G., Wong, K., et al., 1999. The mating-type and pathogenicity locus of the fungus Ustilago hordei spans a 500-kb region.
Proc. Natl. Acad. Sci. U. S. A. 96:15026-15031.
Lovely, C.B., Aulakh, K.B., and Perlin, M.H., 2011. Role of Hsl7 in morphology
and pathogenicity and its interaction with other signaling components in the plant
pathogen Ustilago maydis. Eukaryot.
Cell 10:869-883.
Topp, C.N., Ruiz-Herrera, J., Martinez-Espinoza, A.D., and Gold, S.E.,
2002. Integration of the gene for carboxin resistance does not impact the Ustilago maydis-maize interaction. Curr.
Microbiol 44:67-70.
Tsukuda, T., Carleton, S., Fotheringham, S., and Holloman, W.K., 1988.
Isolation and characterization of an autonomously replicating sequence from Ustilago maydis. Mol. Cell. Biol.
8:3703-3709.
Verma, A., Kapros, T., and Waterborg, J.H., 2011. Identification of a
replication-independent replacement histone H3 in the basidiomycete Ustilago maydis. J. Biol. Chem. 286:25790-25800.
Verma, A., Kapros, T., and
Waterborg, J.H., 2016. Generation of an Ustilago maydis ade2 mutant. Submitted.
Wahl, R., Wippel, K., Goos, S.,
et al., 2010. A novel high-affinity sucrose transporter is required for
virulence of the plant pathogen Ustilago
maydis. PLoS biology 8:e1000303.
Zameitat, E., Freymark, G.,
Dietz, C.D., et al., 2007. Functional expression of human dihydroorotate dehydrogenase
(DHODH) in pyr4 mutants of Ustilago
maydis allows target validation of DHODH inhibitors in vivo. Appl. Environ. Microbiol. 73:3371-3379.