Segregation, Linkage and Epistasis in Neurospora crassa
JANE YEADON
Contents
Topic���������������������������������������������������������������������������� �� ��������������� ����Section
Useful
sources of information��������������������������������������������������� �� ��������������� �3
Notes�������������������������������������������������������������������������������������� �� ��������������� �3
Preparation
for the classes�������������������������������������������������������� �� ��������������� �5
Week
1 (tutorial) ��������������������������������������������������������������������� ������������������ �6
PART
A - Linkage and segregation in Neurospora
crassa
Week
2������� ��������������������������������������������������������������������������� ������������������ �6
Week
3������� ��������������������������������������������������������������������������� ������������������ �7
Week
4������� ��������������������������������������������������������������������������� ������������������ �7
Week
5������� ��������������������������������������������������������������������������� ������������������ �8
Biochemical
assay (glutamate dehydrogenase) �������������������������� ������������������ �8
PART
B - Epistasis in the carotenoid pathway of Neurospora
crassa
Week
3������� ��������������������������������������������������������������������������� ������������������ 10
Week
4������� ��������������������������������������������������������������������������� ������������������ 11
Week
5������� ��������������������������������������������������������������������������� ������������������ 11
Week
6 (tutorial) ��������������������������������������������������������������������� ������������������ 11
Data
analysis���������������������������������������������������������������������������� � ���������������� 12�����
Useful sources
of information
Your textbook, "An Introduction to Genetic Analysis"; the life cycle of Neurospora crassa is in chapter 3, "Chromosome Theory of Inheritance".
http://www.fgsc.net/2000compendium/NewCompend.html
-This is the
http://www-genome.wi.mit.edu/annotation/fungi/neurospora/
-This is the Neurospora genome site
http://vector.cshl.org/dnaftb/
-A teach yourself Genetics site
Have a look around, read and educate yourselves!
Notes
Neurospora crassa - a (very beautiful!) haploid filamentous fungus with seven chromosomes known as LGI-LGVII (Linkage Groups). Neurospora has two mating types, A and a, and cannot self-fertilise. Mating occurs when both mating types grow together under conditions where nutrients (mainly nitrogen) are limited, producing haploid ascospores (see picture on front section) after 4-8 weeks, depending on the strains involved. Never call Neurospora yeast; yeasts are single-celled eukaryotes with simplified lifestyles and genomes. Neurospora is a much more complex multicellular organism, and of course much prettier!
Conidia - asexual multinuclear spores that look like fine talcum powder. They are killed by heat but survive freezing well.
Spores - sexual uninuclear ascospores. They need heat to germinate but are killed by freezing.
Hyphae - segmented multicellular filaments; the vegetative form of Neurospora. Conidia bud from hyphae.
Genetic nomenclature
his+ - a gene whose product is an enzyme involved in the histidine biosynthesis pathway
his - the gene has a
mutation and produces a non-functional enzyme so the strain requires histidine
to grow
cot-1 - a colonial temperature-sensitive mutation. Wild type (cot+) strains produce long unbranched hyphae when grown on sucrose at 25�C and at 34�C. cot-1 strains are colonial at 34�C because the mutation results in frequent hyphal branching, but wild type in morphology at 25�C.
SGF - sorbose glucose fructose. This carbon source restricts hyphal extension, and results in colonial morphology.
Supplements
his - L-histidine
ad - adenosine
ala - L-alanine
arg - L-arginine
gly - glycine
Sterile technique
Sterile technique means different things in different circumstances.
When you are working with DNA, it means avoiding contamination of your samples with enzymes that may digest your DNA; your skin is loaded with such enzymes, so it helps to wear disposable gloves. When you are doing a PCR (polymerase chain reaction), it also means avoiding contaminating your reactions with DNA, particularly DNA that may amplify as well as your template. Once again, it helps to wear gloves, but you also need to work in a clean environment (for example, a laminar flow cabinet), ensure that all your solutions are DNA-free and structure your work in order to minimise cross-contamination of your reactions.
When you are working with microorganisms, you need to avoid contaminating your cultures with organisms that you do not want to grow. When culturing bacteria or fungi, you do not want inadvertently to grow bacteria that live on your skin or inside your body or fungi that are floating in the air. To this purpose, avoid touching any surface that will contact the culture medium, for example, the rim of a culture bottle or a pipette tip used to transfer a suspension of organisms. Do not cough or sneeze near your work! Always work close to a flame to minimise airborne contamination. After you remove a lid and before you replace it, pass the rim of the bottle or flask through the flame to kill any organisms that may be on it.
When working with fungi that propagate themselves by means of small, light asexual spores (such as Neurospora), you also need to remember that your organism is itself capable of being an airborne contaminant. Firstly, I will assure you that Neurospora is not dangerous to you. It is non-toxic, in fact is edible and totally non-pathogenic. So you do not need to protect yourselves from this organism, but you do need to protect your work and that of others from it. For example, if you are trying to assign genotypes to your cultures by determining their phenotypes with respect to nutritional requirements, as you will do in this practical, you want to be sure that the culture you have grown is the one you intended to grow. As well as confusing your own results and possibly those of your laboratory classmates, allowing conidia to escape may result in contamination of cultures of other people in the building. We do not wish this to happen, so you must take special precautions.
Whenever you open any Neurospora culture that includes conidia (slope cultures or mature crosses), do so as close to a flame as you can manage without setting fire to the bung. Remove the bung without rush, with the mouth of the tube directed towards the flame. In this way, any conidia that swirl out of the tube as you disturb the air whilst removing the bung are likely to be incinerated immediately.
When you need to obtain a small number of spores from a Neurospora cross, open the tube as described previously. You then need to remove as many as possible of the conidia at the top of the culture prior to obtaining the spores. Use a hot loop and scrape up conidia, then transfer them directly from the mouth of the tube into the flame for incineration.
Please be careful, but do not panic; we do not expect perfection.
Preparation
for the classes
Questions to be
attempted prior to week 1:
Humans are diploid organisms, with 46 chromosomes, two copies of each chromosome per person. A single gene determines blue versus brown eyes in humans. In other words, the blue-eye gene is an allele of the brown eye gene. The brown allele is dominant to the blue allele, meaning that you need two copies of the blue allele to have blue eyes.
1. If you and your spouse have blue eyes, what colour eyes will your children have?
2. If one of you has blue eyes and the other has brown, what colour eyes will your children have?
What do you need to know to answer this question?
The gene for albinism (pale skin and eyes, white hair) in humans is recessive, so you need two copies of this gene to be an albino.
3. If you marry an albino, what proportion of your children will be albino? What do you need to know to answer this question?
4. If you have blue eyes and you marry an albino, what colour eyes will your children have? What do you need to know to answer this question?
Imagine that humans are haploid (ignore the technical details!). Now answer questions 1-4 again (call them 5-8, to save confusion). Which questions were easier?
Questions to be
attempted prior to week 2:�
1. How many spores will you put on each of the A plates?
2. How many on each of the B plates?
3. Why do you think these two dilutions are necessary?
Questions to be
attempted prior to week 3:
1. How many spores of the albino by yellow cross will you put on each plate?
2. Why are the plates incubated at 25�C?
3. Why are the progeny of the three-point-linkage cross to be picked from fully supplemented plates?
Prior to week 4:
Explain the set of selective plates onto which you will patch your progeny of the three-point-linkage cross. Remember that am mutants will grow without alanine (slowly) but will not grow at all in the presence of glycine.
Prior to week 5:
Generate a table with all the genotypes that may be present in progeny of the three-point-linkage cross. This is VERY important; if you do not do this you may have too little time to complete the biochemical assay.
Prior to week 6:
Review (or learn) how c2 tests are performed and what statistical significance and degrees of freedom (df) mean. Think about what is happening in meiosis and how this affects the numbers of progeny of each type. Attempt at least one c2 test.
Discussion questions: 2nd Year Biotechnology Genetics Practical
1a. You have a cross between 2 strains of Neurospora crassa, one of which requires adenine to grow and the
other grows on minimal medium. You plate approximately 1000 spores on medium
with adenine added and count 990 colonies. If you plate the same number of
spores on minimal medium, how many colonies do you expect? About 500� Why? You asssume that there is a single allelic
difference segregating in this cross. One parent has the ad+ allele
and the other the ad mutant allele. If the alleles are segregating normally,
every spore from this cross has an equal chance of getting either allele. Thus
half of the progeny will grow without adenine.
b. When you plate the spores on minimal medium, only 250 colonies
grow. How could this be? The
adenine-requiring mutant could have mutations in two unlinked genes. In ither
words it is a cross between ad-A ad-B�
and ad-A+ ad-B+ strains (making up gene names at
random!) How could you test your hypothesis? Cross the adenine-requiring progeny to a wild-type strain. 2/3 of the
crosses should yield 50% wild type progeny (ad-A ad-B+ or ad-A+
ad-B crossed to ad-A+ ad-B+) and 1/3 only 25% as above
(ad-A ad-B crossed to ad-A+ ad-B+).
2. This time your cross is between a strain that requires lysine and another that requires arginine. You plate the same amount of spore suspension on plates with adenine and arginine, just adenine, just arginine and minimal medium.
Fully supplemented������������������ 1000
lysine��������������������������������������� 500
arginine������������������������ 500
minimal������������������������ 250
What can you conclude? The genes determining lysine and arginine
requirements are unlinked. This means either:
they are on different chromosomes and thus are assorting independently or: they are so far apart on the same
chromosome that they behave as if they are on different chromsomes. In this
case, a crossover would occur between the two genes in virtually all meioses.
3. A cross between the same lysine-requiring strain and one that requires histidine.
Fully supplemented������������������ 1000
lysine��������������������������������������� 500
histidine����������������������������������� 500
minimal������������������������ �50
How can you explain this difference? The genes determining lysine and histidine requirements are linked. This means that they are on the same chromosome. A crossover occurs between the two genes relatively infrequently. If the parental genotypes are lys his+ and lys+ his, 50 out of 1000 progeny are lys+ his+, a frequency of 5%. Since we can assume that an additional 50 progeny would be lys his, the frequency of crossing over between lys and his loci is 10%. The genetic distance between these loci is thus 10 map units (mu), or 10 centiMorgans (cM).
4. A colleague gives you a Neurospora culture of a very interesting lilac colour, saying that it turned up in a mutagenesis experiment. The culture requires adenine and you are known to have an interest in genes involved in adenine biosynthesis.
You cross the lilac strain to a
wild type strain (orange, adenine-independent). You plate spores on media with
and without adenine. As expected, only half of the spores grow on minimal
medium, whereas more than 98% germinate on adenine-supplemented medium. Shows that there is only one gene with a
mutant allele determining adenine requirement segregating in this cross.
You pick 64 colonies from the supplemented plates to supplemented slopes and another 64 from minimal plates to minimal slopes.
����������������������� orange������������� lilac����������������� white��������������� brown/purple
supplemented�� ��� 19��������������� � 16����������������� � 14����������������������������� 15
minimal ��� 34��������������� �� -������������������� � 30����������������������������� -
You patch all the cultures from the supplemented slopes onto medium with and without adenine. All the lilac and brown cultures require adenine and none of the white or orange cultures do. What do you think is happening?
Clearly the pigment giving a lilac or purple colour is closely linked
to (or determined by) adenine requirement. When you remove the complication of
the adenine requirement, you see that white and orange are segregating
normally. Thus the cross must be of the nature ad wht� x ad+ wht+. The lilac
colour results from the purple pigment due to adenine requirement in a strain
in which the orange carotenoid pigments are not being made. Brown/purple
strains are ad wht+, orange strains are ad+ wht+
and white are ad+ wht. Additionally, you can tell that ad and wht
are unlinked as all four genotypes are present in equal numbers.
Week 1
Tutorial/pre-lab (2-3 hours) on haploid genetics, the Neurospora life cycle and the practical assignment. The display will include Neurospora strains of various genotypes, mature crosses and plates with colonies both cot-1 and cot+ (perhaps the plates will be replaced by photographs?). his, arg and wild-type strains are orange, al-2 is white, and ylo-1 yellow. ad mutants are purple/brown, due to a deep red pigment, unrelated to carotenoids, that accumulates when adenine is limiting. A series of questions related to haploid genetics will be discussed informally (previous section).
PART A - Linkage and
segregation in Neurospora crassa
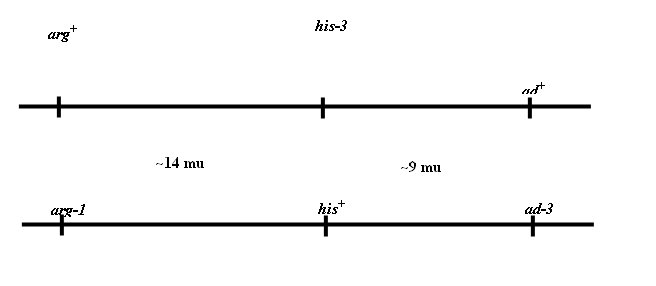
The cross: ad+ am
arg+ his A x ad am+
arg his+ a
Both strains are cot-1.
Week 2
Extraction of progeny from the linkage cross: A pre-prepared spore suspension (14,000 spores/ml) from this cross is available. Mix the suspension thoroughly by vortexing, then immediately transfer 1 ml of suspension into 19 ml layer agar (bottle A) at 60�C. Vortex the suspension in bottle A thoroughly, then put 1 ml into another 19 ml layer agar (a 1/20 dilution) to give bottle B. Replace the bottles in the 60�C water bath and incubate for 60 minutes (actually 45-70 minutes; less than 45 minutes and not all the conidia will be killed, more than 70 minutes and the spores will begin to die).
Label your plates on the lids (so the writing will not interfere with counting colonies next week) with your names and "A" or "B". Keep the plates at 34�C until you need them; this will ensure that the layer agar does not set too quickly.
Plating procedure: The plating procedure should be performed as quickly as possible but sterile technique should be used throughout. The bottle containing the spores in molten agar should be mixed thoroughly by vortexing before each pipetting operation, as you need each aliquot from the same bottle to contain approximately the same number of spores.
Pipette 3 ml of spore suspension from bottle A onto each of the ad ala (6), arg ala (7) and ala (8) SGF plates provided and tilt plates gently to distribute the agar evenly.� Remember to mix suspension between each pipetting operation.� Use a fresh tip for each 3 ml aliquot.
Pipette 3 ml of spore suspension from bottle B onto his ad arg ala (1), his ad arg gly (2), ad arg ala (3), ad arg gly (4) and his ala (5) SGF plates.
When top layer agar is set, invert the plates so the lids are on the bench, form into two piles and wrap each with a rubber band.
Incubate the plates at 34�C for 3 days. Then keep them at 4�C until the next class.
Week 3
Recording the numbers of surviving progeny on each type of selective plate: Count the colonies on each plate and record.
Table 1:
|
Bottle |
Plate # |
Plate supplement(s) |
Number of �colonies |
|
B |
1 |
his ad arg ala |
|
|
B |
2 |
his ad arg gly |
|
|
B |
3 |
ad arg ala |
|
|
B |
4 |
ad arg gly |
|
|
B |
5 |
his ala |
|
|
A |
6 |
ad ala |
|
|
A |
7 |
arg ala |
|
|
A |
8 |
ala |
|
You expect the numbers of colonies on plates 1, 2 and 3 to be in a ratio of 2:1:1 (why?). Perform a c2 test to assess if this is the case. If your c2 value is significant, let your demonstrator know, as your data may not be suitable for generation of combined class data.
Subculture of unselected progeny for additional estimates of linkage: this is particularly useful if you have not succeeded in plating the same numbers of spores on each of your B plates (shown if your c2 test 1(a) is significant; Data Analysis section).
Pick 32 progeny at random from the fully supplemented plates (his ad arg ala; #1) into fully supplemented slopes, labeled 1-32. This is done with the plate under the dissecting microscope, so you can see the colonies clearly. Heat a spade-ended needle until red, cool then use the needle to cut a rectangular section of the colony. Use the needle to lift the rectangle away from the plate. Remove the cotton plug from a slope, flame the top of the tube and place the rectangle with the colony onto the agar slope. Flame the top of the tube and replace the plug. Incubate the slopes at 25�C until the next class.
Week 4
Pooled class results: Your demonstrator will supply you with the class data from week 3 for statistical analysis. This is the summation of plate counts from Table 1, not including data-sets in which
(i) The preliminary c2 value was significant�
(ii) Other errors appear to have occurred
Subculturing of each of the unselected progeny on selective media: Patch conidia from each of your 32 cultures (16 per student) to SGF plates:
his ad arg ala (1), his ad arg gly (2), ad arg ala (3), his ad ala (4), his arg ala (5).
You will need to draw a grid of 4 x 4 squares on each plate and label each square 1-16 or 17-32. Using sterile technique, pour some sterile water into an empty petri dish. Heat a wire loop until it is red and dip it into the water. Remove the cotton plug from a slope culture, flame the top of the tube. Use the loop to mix some conidia from the slope with the water in the loop and replace the plug. Touch the loop to the surface of the plates in the desired positions. Remember to heat the loop to kill conidia when you are finished.
Incubate the plates at 25�C for 2 days, then keep at 4�C until the next class.
Inoculate two of the 32 cultures
into his ad arg ala liquid medium (50 ml in�
a 250 ml flask). Label each flask with your name and the isolate number
of the culture (1-32). Incubate the flasks at 25�C with shaking for 48 hours,
then keep at 4�C until the next class.
Week 5
Determination of the genotypes of each of the unselected progeny: Record which cultures grew on which selective plates and determine their genotypes. Record these on the table (demonstrator's copy provided on section 15) generated prior to the class. The table below is a reminder of the purpose of each selective plate.
Table 2:
|
Medium |
Lack of growth means... |
|
his ad arg ala |
experimental error! |
|
ad arg ala |
his |
|
his arg ala |
ad |
|
his ad ala |
arg |
|
his ad arg gly |
am |
Pooled class results: Your demonstrator will pool the data from the unselected progeny whose genotypes you have determined by patching onto selective media. This combined data will be used for statistical analysis.
Biochemical assay (glutamate dehydrogenase)
Neurospora am mutants do not make NADP-dependent glutamate dehydrogenase (GDH). The reaction catalysed by the product of the am gene is:
glutamate + NADP+ �� a-ketoglutarate + NH3 + NADPH + H+
NADPH absorbs light at 340nm. We can therefore use an increase in absorption at 340nm as a measure of the activity of the am gene in each of the two cultures grown in liquid medium.
Since you have now determined the genotypes of all 32 unselected progeny cultures, you should know whether the cultures you grew in liquid medium are both am, both am+ or (if you are lucky) one of each. If you are not lucky, you may swap with someone else so that you each have one of each genotype. If this is not possible, obtain additional results at the end of the class so that you have data for both am and am+ assays.
Preparation of crude protein extract: The mortar should be on ice to keep it cool. Pour most of the medium from one flask. Flame your forceps, cool, then use to pull the lump of mycelium out of the flask and lie it on a wad of paper towel. Use the towel to remove as much of the moisture as possible. Place your damp mycelium into the mortar. Add about half a teaspoon of sand and grind into a paste. Add 5 ml 100mM sodium phosphate buffer, 1mM EDTA (pH 7.4) and grind until homogeneous. Pour into 10 ml centrifuge tube, avoiding most of the sand. Store on ice and repeat with the other culture (clean mortar and pestle with alcohol in between). Ensure that both tubes contain the same volume of extract before centrifugation and, if necessary, top up with sodium phosphate EDTA buffer.
Centrifuge the sand/extract mixture (4000 rpm/10 min/4�C). Remove as much as possible (3-4 ml) of the supernatant into clean tubes. Store on ice.
GDH Assay: Assay ONE culture at a time.
Assay mixture:� 1 ml extract
����������������������������������� 1 ml 0.2% NADP (in 100mM Tris pH 8.5)
����������������������������������� 7 ml 100mM Tris pH 8.5
Mix thoroughly by inversion. Put 2.7 ml of the mixture into each of three cuvettes. To the first, add 0.3 ml 100mM Tris pH 8.5. Mix by inversion (use a small square of parafilm between the cuvette and your finger). This is your blank. To each of the other two cuvettes, add 0.3 ml of the substrate monosodium glutamate (2.5M in 100mM Tris pH 8.5) and mix as before. Start the timer as soon as you mix, as this starts the reaction. Record the OD340 of each of your assay mixtures every minute for a minimum of five minutes. Zero your blank before each pair of readings. Repeat once more for the same culture, then do it all over again twice for the other culture.
Plot a graph of OD340 against time. Can you tell which cultures are am and which ones are am+?
Do you think that GDH is the only source of NADPH in the biochemical assay? Why do you have to zero your blank before each reading?
PART B - Epistasis in the
carotenoid pathway of Neurospora crassa
al-2: albino-2
Location - IR. Carotenoids absent or abnormal. Defective in
phytoene synthetase, with a lesion between prephytoene pyrophosphate and
phytoene.
ylo-1: yellow-1
Location - VIL Yellow carotenoids. Affects synthesis of
neurosporaxanthin (4'-apo-beta'-caroten-4'-oic acid). Lesion probably
involves the conversion of lycopene to 3,4-dehydrolycopene or the
conversion of either torulene or gamma-carotene to neurosporaxanthin.

Figure 1. Biosynthetic pathway for carotenoids

|
al-2: albino-2 Location - IR. Carotenoids absent or abnormal. Defective in
phytoene synthetase, with a lesion between prephytoene pyrophosphate and
phytoene. ylo-1: yellow-1 Location - VIL Yellow carotenoids. Affects synthesis of
neurosporaxanthin (4'-apo-beta'-caroten-4'-oic acid). Lesion probably
involves the conversion of lycopene to 3,4-dehydrolycopene or the
conversion of either torulene or gamma-carotene to neurosporaxanthin. |
|
Figure 1. Biosynthetic pathway for carotenoids |
The cross: al-2 a (white) x ylo-1 A (yellow)
Both strains are cot+.
Week 3
Extraction�
of progeny from the al-2 x ylo-1 cross:
Flame a microscope slide and a
scalpel.� Allow to cool and cut a strip
(~0.75 cm x 3 cm ) of 2% agar from a petri dish and place on the slide.
Remove the plug of conidia above the paper insert
in the crossing tube with a flame sterilised loop.� Care should be taken with this to ensure that
you do not spill conidia on the bench-top.
Flame a wire loop, cool in sterile water and touch
the wet loop onto the black ascospores adhering to the wall of the tube (not
the paper insert).� Streak the loop
gently along the agar block. Care should be taken not to transfer conidia
(small and colourless under the microscope) or perithecia (black and 0.6 mm
long) to the agar block.� Ascospores are
black, shaped like footballs, and about 0.03 mm long.
Using a dissecting microscope, count the number of
black ascospores on the block
(non-pigmented spores are immature and will not germinate).� You need a number in the range 100-120.� If there are not enough spores on the block
transfer more with the flamed sterile loop.
Record the number of spores on the block and
transfer it to a bottle containing 10 ml of molten agar and keep in a 60�C water bath.�
Label the bottle with your name and date and return to the water bath as
quickly as possible so the agar will not solidify.� Incubate for 60 minutes.
Label the two remaining minimal SGF plates provided with your name and the cross. Keep
at 34�C until you are ready
to plate your spores.
Plating procedure: During the plating procedure, sterile technique should be used throughout and the bottle containing the spores in molten agar should be mixed thoroughly by vortexing before each pipetting operation.
Pipette 3 ml of spore suspension onto each of the two labelled minimal SGF plates and tilt plates gently to distribute the agar evenly.� Remember to mix suspension between each pipetting operation.� Use a fresh tip for each 3 ml aliquot. Incubate plates at 25�C for 48 hours, then keep at 4�C until the next class.
Week 4
Pick 8 progeny from the minimal plates to minimal slopes, labeled 1-8. Incubate the slopes at 25�C until the next class.
Week 5
Score cultures: Count cultures of each colour (white, yellow and orange). Pool data.
Table 3:
Colour
|
Number
|
white |
|
orange |
|
yellow |
|
Why are some of the progeny of a cross between a yellow and an white strain orange?
What proportion of progeny are white? What proportion are yellow? What proportion are orange? Explain.
If you had a strain that was al-2 ad, what colour do you think it would be and why?
Week 6
Discuss analysis of data and explanations (large group tutorial).