Acriflavine Resistant (acr)
Acetate Utilization (acu)
Adenine (ad)
Albino (al)
Amination (am)
A/a: mating type alleles
IL. Between un-3 (0.04 to 0.1%) and un-16 ( < 1%) (488,
758; D. D. Perkins,
unpublished
data). (609)
Opposite mating types are essential for a complex of events associated with sexual reproduction and morphogenesis: attraction of trichogyne to cells of opposite mating type (39, 93); pickup and transport of the nucleus to the ascogonium; growth and development of the perithecium; proliferation of heterokaryotic ascogenous hyphae; conjugate nuclear divisions in precrozier and crozier cells; karyogamy.
Mating type alleles also act as vegetative incompatibility genes during the vegetative phase. A+a combinations are unable to form stable heterokaryons (66, 384, 830, 914). Vegetative fusion is usually followed by cell death (384), but some A+a heterokaryons grow slowly (252, 412, 422). Heterozygous A/a duplications are highly abnormal, with inhibited growth and spider-like morphology (761. 804). Incompatibility in heterokaryons or duplications is relieved by spontaneous deletion of either allele (252, 756). Vegetative incompatibility is not expressed during the sexual phase after fertilization. Both manifestations of vegetative incompatibility are suppressed by tol, but sexual compatibility is not affected (755). The vegetative incompatibility is normally suppressed in N. tetrasperma (668) and N. sitophila (674, 804). Extensive efforts have failed to separate the sexual and vegetative traits by genetic recombination (758). Null mutants selected by loss of vegetative incompatibility usually lose both sexual and vegetative functions simultaneously (one exception), and both functions are usually restored simultaneously in revertants selected for restoration of fertility (one null mutant gives atypical revertants) (252, 411, 412).
Only two mating type alleles, A and a, are known. These are apparently homologous throughout the genus Neurospora (820) and perhaps in related genera (770). Nothing is known about the genetics of the five true-homothallic species of Neurospora, which closely resemble N. crassa in karyotype and meiotic behavior, including the fusion of two haploid nuclei in the penultimate cell of the crozier to form the zygote nucleus (855). In the early literature, A was called + (plus) or A, and a was called - (minus) or B (e.g., reference 286). The locus may also be designated mt, mating type (e.g., reference 808), and is usually referred to as mt in the present paper.
aaf: acetylaminofluorine requirement
Data said to be consistent with one gene.
Complex phenotype. Alternative requirements: 2-acetylaminofluorine, certain azo dyes, or certain single amino acids. Cold sensitive. Originated among progeny of a rib-1 strain that had become tolerant to 2-acetylaminofluorine. Abstract only: 1069.
ac: acetate
Changed to ace.
ace: acetate
Acetate mutants ace-1 through ace-7 are auxotrophs that grow on
0.3% sodium
acetate,
as do suc mutants (which often grow better on acetate than on succinate). A
carbon
source
is also needed. Most ace mutants grow better when the carbon source is
maltose rather
than sucrose (578). Acetate mutants, except ace-1 and ace-5, can
grow on Tweens as the
sole carbon source (S. Brody, personal communication). The mutants differ in their
ability
to grow on complex media. Unlinked genes ace-2, -3, and -4 are
involved with the
pyruvate
dehydrogenase
complex (769). A separate set of acetate mutations called ac-1, -2, -3, 4, and
-5 (1034)
were
not mapped and are not available for testing for allelism with ace-1 through
ace-7.
General
reference: 578.
ace-1: acetate-1
IIR. Right of un-20 (15%) and ure-3 (14%). Left of fl
(2 to 11%) (47, PB).
Requires acetate. Poor growth on complex complete medium. Grows well on acetate (0.1 %) aided by ethanol (0.5%) (E.L. Tatum and L. Garnjobst, cited in reference 47; PB). Ascospore maturation and germination are slow. Germination is best on sucrose minimal medium with yeast extract and ethanol (L. Garnjobst, personal communication). Not the same as ac-1 of reference 1034, which was lost. Called ac.
ace-2: acetate-2
IIIR. Right of pro-1 (1 to 9%). Left of com (5%) and
ad-4 (4 to 7%) (578).
(812)
Requires acetate. Will not use succinate or ethanol (290). Lacks pyruvate dehydrogenase complex activity (769). Good growth on complex complete medium (578). Not the same as ac-2 of reference 1034.
ace-3: acetate-3
IR. Right of In(OY323) and, hence, of lys-3 (0.3 to 1%). Left of
nic-1 (0.2%).
Included
in duplications from In(OY323) x In(NMI76). (2, 57, 578,
907)
Requires acetate. Lacks pyruvate dehydrogenase complex activity (769). Poor growth on complex complete medium (578). Conidiation best at 25 C, not 34°C (D.D. Perkins, unpublished data). Not the same as ac-3 of reference 1034.
ace-4: acetate-4
IVL. Between cys-10 (19 to 33%) and fi (10 to 17%)
(578).
Requires acetate. Grows on complex complete medium (578). Lacks pyruvate dehydrogenase complex activity. Lipoate acetyltransferase fails to aggregate to form the core of the pyruvate dehydrogenase complex. As a result, there is high activity of the free components pyruvate dehydrogenase and lipoamide reductase (769). Not the same as ac-4 of reference 1034.
ace-5: acetate-5
VR. Between gul-1 (< 1%) and ure-1 (< 1%) (577
578)
Requires acetate. Poor growth on complex complete medium (578). Not the same as ac-5 of reference 1034.
ace-6: acetate-6
See suc.
ace-7: acetate-7
IR. Between nic-2 (4 to 7%) and cr-1 (1 to 3%) (578).
Requires acetate. Good growth on complex complete medium. Unable to use xylose as a carbon source; resembles suc mutants and differs from the wild type and all other ace mutants in this respect. Normal pyruvate dehydrogenase and pyruvate carboxylase activities. (578)
acon(-1): aconidiate
See fl.
acon-2: aconidiate-2
IIIR. Linked to vel (6%) and tyr-1 (9 to 14%), probably between
them (648, PB).
Macroconidiation defective. Allele RS91 is heat sensitive, with macroconidiation blocked at 34°C. Some conidia are formed at 25 C, but growth is subnormal (648). Homozygous fertile (PB).
acon-3: aconidiate-3
IVL. Between cys-10 (1 to 6%) and cut (33%) (PB). Report of
VIL linkage not
confirmed.
Macroconidiation blocked (648). Female sterile. Some conidia have been observed low in slants at 25°C (PB).
acpi: acetate permease (inducible)
Not mapped. Not allelic with other acetate utilization (acu) mutations.
Lacks inducible acetate transport system. (864, 866)
acr-1: acriflavine resistant-1
IL. Linked to mt (8 to 12%) (498).
Low-level resistance to acriflavine (2 µg /ml) was found for wild-type STA4 compared with that for wild-type Pa, which is more sensitive (498). Difficult to score.
acr-2: acriflavine resistant-2
III. Linked to thi-4 (0/286). Left of sc (3 to 6%) and
spg (1 to 11%) (498,
816). acr-2 has
been shown left of the centromere on published maps but without direct evidence.
acr-2
and
trp-1 (on IIIR) cosegregated at the second division in 1 of 13 asci (H.B. Howe,
Jr.,
personal
communication), which would favor a right arm location.
Resistant to acriflavine (494, 495); also resistant to 3-amino-1,2,4-triazole (seven alleles tested) (494). Resistance is probably dominant (heterokaryon tests) (498). Not resistant to malachite green. An excellent stable marker, fully fertile, with unambiguous scoring. Sizable inocula should be used to avoid false-negative tests. Use acriflavine at 50 µg /ml in minimal agar medium (816) (higher concentrations may be used) and aminotriazole at 0.5 mg/ml; both added before autoclaving.
acr-3: acriflavine resistant-3
IL. Between un-16 (1 to 5%) and suc (1 to 5%). Probably right of
ta (816; PB).
(498)
Resistant to acriflavine and to malachite green (three alleles tested). Not resistant to 3-aminotriazole. Resistance is probably dominant (heterokaryon tests) (498). Scoring is clear-cut with uniform inocula of appropriate size. False-negative or false-positive scoring may result if test inocula are too small or too large. May show delayed resistance: read tests at 2 and 4 days, 34°C. Use acriflavine at 10 µg /ml in minimal agar at 34°C (816) and malachite green at 2 µg /ml (498).
acr-4: acriflavine resistant-4
I. Linked to mt and acr-3 (5%) (499).
Resistant to acriflavine (50 µg /ml) when acr-4 is combined with morphological mutation shg (499).
acr-5: acriflavine resistant-5
I or II. Linked to T(IR;IIR)4637 al-1 (499).
Resistant to acriflavine (50 µg /ml) when acr-5 is combined with linked morphological mutation mo(KH161) (499).
acr-6: acriflavine resistant-6
IIIR. Linked to shg (0/368) (499).
Resistant to acriflavine (50 µg /ml). Originated in shg and not separated by recombination. Strain of origin is acriflavine sensitive. (499)
acr-7: acriflavine resistant-7
IIIL. Left of sc (12 to 14%). Linked to thi-4 (7%) (PB). Right
of r(Sk-2)-1 (7%)
(B.C.
Turner, personal communication). Report of VI linkage in reference 818, is incorrect.
Resistant to acriflavine (50 µg /ml). Not cross-resistant to 3-aminotriazole or malachite green. Several acr-7 strains have become female infertile after vegetative transfer. (PB)
act: actidione resistant
Changed to cyh (cycloheximide resistant) (807).
acu: acetate utilization
For a diagram of the pathway, see p. 304 in reference 343. Scored on minimal
medium
without sugar, using ammonium acetate (3 mg/ml) as the carbon source. The wild type
shows sparse but clearly positive growth in contrast to clear blanks for acu mutants.
Selectable by inositol-less death on acetate medium. acu-1, acu-5, acu-6, and
acu-7 do
not
behave as respiratory mutants in tetrazolium overlay tests on acetate medium (310).
acu-1: acetate utilization-1
VR. Right of asn (21%) (349).
Unable to use acetate as a carbon source (349, 350). Selected by inositol-less death on acetate medium.
acu-2: acetate utilization-2
IVR. Between leu-2 (11%) and pan-1 (6%) (349).
Unable to use acetate as a carbon source. Reduced level of oxoglutarate dehydrogenase. Poor recovery from ascospores (349, 350).
acu-3: acetate utilization-3
VR. Between inl (7%) and asn (20%) (590). (349)
Unable to use acetate as a carbon source (349, 350). Affects isocitrate lyase (350, 590). Some revertants produce temperature-sensitive enzyme (590).
acu-4: acetate utilization-4
IR. Right of arg-1 (5/29) (349).
Unable to use acetate as a carbon source (349, 350).
acu-5: acetate utilization-5
II. Linked to arg-5 (6%) and aro-3 (7%) (349).
Unable to use acetate as a carbon source. Affects acetyl coenzyme A synthetase (350).
acu-6: acetate utilization-6
VIL. Left of cys-1 (3%) (76, 349).
Unable to use acetate as a carbon source (349). Structural gene for phosphoenolpyruvate carboxykinase (76, 350). Strains with some complementing alleles possess protein that is electrophoretically similar to the enzyme; a temperature-sensitive partial revertant allele specifying an abnormally thermolabile enzyme maps at the original locus (76). Interallelic complementation (349).
acu-7: acetate utilization-7
IIIR. Linked to dow (0/72) (PB).
Unable to use acetate as a carbon source. Poor recovery from ascospores (~25%) (349). Reduced level of oxoglutarate dehydrogenase (350).
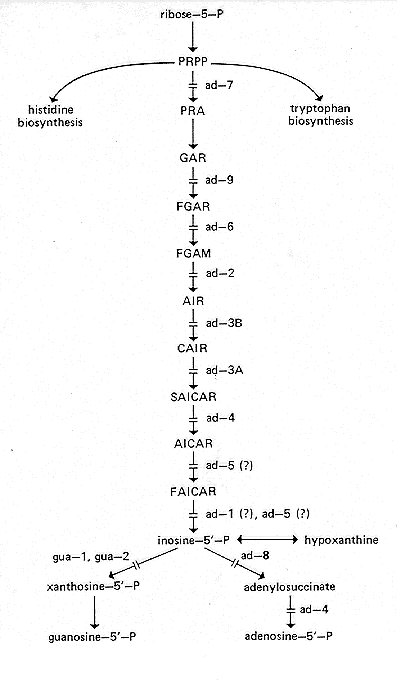
ad: adenine
For the purine biosynthetic pathway, see Fig. 8. (For the
abbreviations used below, see
the legend to Fig. 8.) For the interrelationship of purine,
histidine, and tryptophan
pathways, see reference 786. The growth of mutants in the terminal (post-AICAR) steps
of the adenine biosynthetic pathway is aided by histidine, which has a sparing effect on
ad-1,
ad-4, and ad-8 (661; M.E. Case, personal communication). Mutants
carrying some ad-5
alleles are aided by histidine; others are inhibited (M.E. Case, personal communication).
Indole may strongly inhibit adenine mutants (595).
Strains carrying mutations at loci ad-3A and ad-3B accumulate purple (red) pigment when adenine is limiting; see the ad-3B entry. Smaller amounts of the pigment may be seen in strains carrying other post-AIR genes such as ad-4 and ad-5 (526, 682). Pigment may be polymerized AIR (786). Mutant genes affecting earlier biosynthetic steps are epistatic to ad-3 and later mutant genes with respect to pigment production (526). ad-3B (and presumably also ad-3A) stocks accumulate spontaneous mutations at other ad loci, which prevent pigment production and improve the growth rate (691).
ad-3B, ad-4, ad-8, and ad-9 mutants have been used to study the effect of histidine on purine pool utilization (786). Regulation of purine catabolism reviewed in reference 642. Regulation of purine biosynthesis (405a, 788). Adenine mutations at the various loci were assigned to complementation groups designated by capital letters (526). The relationships of most of these groups to steps of the biosynthetic pathway are given in Fig. 10 of reference 120.
FIG. 8. Purine biosynthetic pathway and sites of action of ad and gua genes (81, 120, 348, 393, 405a, 511, 525). PRPP, 5- phosphoribosyl pyrophosphate; PRA, 5-phosphoribosylamine-, GAR, 5'- phosphoribosyl-glycineamide; FGAR, 5'-phosphoribosyl-formylglycineamide; FGAM, 5'-phosphoribosyl-formylglycineamidine; AIR, 5'-phosphoribosyl-5-aminoimidazole; CAIR, 5'-phosphoribosyl-5- aminoimidazole-4-carboxylate; SAICAR, 5'-phosphoribosyl-5- aminoimidazole-4-N-succinocarboxamide; AICAR, 5'-phosphoribosyl-5- aminoimidazole-4-carboxamide; FAICAR. 5'-phosphoribosyl-5- formamidoimidazole-4-carboxamide.
ad-1: adenine-1
VIL. Right of ylo-1 (6%). Left of the centromere (< 1 to 2%),
T(AR209), and rib-1
(3
to 5%) (1102, 1012). (482)
Uses adenine or hypoxanthine (682, 824). Accumulates AICAR (81, 904) and SAICAR (81). May affect inosine 5'-monophosphate cyclohydrolase (902, 904) (Fig. 8). Used to study purine transport (903 and references therein). Ascospores are white in homozygous ad-1 x ad-1 crosses, and ad-1 ascospores may be white in heterozygous crosses (D.D. Perkins, unpublished data). Called complementation group M.
ad-2: adenine-2
IIIR. Between thi-2 (1%) and trp-1 (1 to 7%) (11, 219).
(482)
Requires adenine or hypoxanthine (682). Controls conversion of phosphoribosylformylglycineamidine to AIR (120) (Fig. 8). Strains carrying allele 70004(t) are heat sensitive (34°C versus 25 C) (682) and osmotic remediable (636). Called complementation group H.
ad-3A: adenine-3A
IR. Between his-3 (1 to 2%) and ad-3B (0.1 to 0.7%) (271).
Right of ure-4 (78).
(482)
Requires adenine or hypoxanthine (682). Blocked in interconversion of CAIR plus aspartate to SAICAR (348) (Fig. 8). Produces purple pigment, permitting direct visual selection (276, 682); see the ad-3B entry. Reduced interallelic fertility (407). No interallelic complementation (267; F.J. de Serres, personal communication). ad-3A and ad-3B are two genetically and functionally distinct loci separated by a short but functionally complex region of unknown but essential function (271, 407). They have been used intensively for quantitative genetic and molecular studies of mutation (for a review, see reference 35). Either forward mutation (e.g., reference 277) or reverse mutation (e.g., reference 772) can be measured precisely; the former is detected visually by purple pigment. Purple pigment has also been used to assess the effect of histidine and tryptophan on purine nucleotide synthesis (786). Alleles N23 and N24 have been used as mutagen testers. N23 reverts with agents that cause base pair substitutions; N24 reverts with agents that cause frameshifts (772). SK(ad-3A) is at or near ad-3A and may be a cryptic ad-3A allele. Does not require adenine. In SK(ad-3A) x ad-3A crosses, the ad-3A progeny die; possibly SK(ad-3A) mutants fail to make enough adenine to support their growth (251). Translocations Y155M64 ad-3A (272; PB) and Y112M15 ad-3A (413) each have one breakpoint that is inseparable from ad-3A. Called complementation group A (264). "A" in the locus symbol does not refer to mating type.
ad-3B: adenine-3B
IR. Between ad-3A (0.1 to 0.7%) and nic-2 (3%) (271).
(482)
Uses adenine or hypoxanthine (682). Blocked in interconversion of AIR to CAIR (348) (Fig. 8). Produces purple pigment, permitting direct visual selection (276, 682). Pigment is secreted with low concentrations of adenine (e.g., 0.1 mM), not with high concentrations (2 mM) (276, 682, 785). Pigment production used to assess effect of histidine and tryptophan on purine nucleotide synthesis (786). Reduced interallelic fertility (264, 407). Complementation maps (268, 274). Relation of mutagens to complementation patterns (269). Mutants with non-polarized complementation patterns on the right side of the complementation map grow on minimal medium if supplied with CO2; other mutants do not respond to CO2, (270). Used extensively for mutagenesis (see ad-3A). Rearrangement T(I- >III)Y112M4i ad-3B, which has a breakpoint inseparable from ad-3B, was the first insertional translocation to be reported for fungi (266). Allele 7-017-0137 shows "fixed instability," mutating to an unstable prototrophic allele (41). Alleles 2-17-126, 12-21-28, and numerous others are supersuppressible (408, 749, 955). Called complementation group B.
ad-4: adenine-4
IIIR. Right of met-8 (1 to 4%) and com (0 to 5%). Left of
leu-1 (1 to 3%) (219,
815).
(482)
Requires adenine. Cannot use hypoxanthine or inosine (661). Growth on adenine (at least for strains carrying allele 44206) is improved by the addition of histidine and still more by histidine plus methionine (661). Structural gene for adenylosuccinase, which controls two reactions in adenine synthesis (393, 1158). (See Fig. 8.) Accumulates a small amount of purple pigment when adenine is limiting (682). Used for the first demonstration of complementation between alleles in vivo (393) (simultaneous with independent demonstration in am) and in vitro (1157). Enzyme in revertants (1158). Used to study purine transport (787). Strains carrying alleles 44206 and 44415 are heat sensitive (34 C versus 25 C) (482, 682) and are osmotic remediable at 30°C (636); enzyme synthesized at 30°C by heat-sensitive strains has altered properties (636). Called complementation group F.
ad-5: adenine-5
IL. Between phe-1 and arg-1 (1%) (816; H.B. Howe, Jr., personal
communication).
(482)
Uses adenine or hypoxanthine (682) (Fig. 8). Accumulates AICAR (81, 904) and SAICAR (81). Some mutants are stimulated by histidine and may not grow on hypoxanthine unless histidine is present; others may be inhibited by histidine (393; M.E. Case, personal communication). Produces some purple pigment, but less than ad-3A and ad-3B mutants (526). Called complementation group J. Evidence, apparently enzymatic, given in reference 120 suggests that some ad-5 mutants lack both AICAR formyltransferase and inosine 5'-monophosphate cyclohydrolase, but apparently other ad-5 mutants lack only the formyltransferase. Indirect evidence (902, 904) suggests that strains carrying ad-5 allele Y112M192 are blocked at the formyltransferase step.
ad-6: adenine-6
IVR. Right of ilv-3 (9%) (579). Left of pan-1 (1 to 2%),
chol-1 (1%), and cot-1 (2 to
6%) (633). (692)
Uses adenine or hypoxanthine (682). Blocked in conversion of phosphoribosylformylglycineamide to phosphoribosylformylglycineamidine (120) (Fig. 8). Inhibited by caffeine in the presence of adenine (1172). Called complementation group 1.
ad-7: adenine-7
VR. Right of cot-2 (4%). Left of ro-4 (4%) and pab-2
(8%). (158, 156).
(687)
Uses adenine or hypoxanthine (682). Lacks phosphoribosylpyrophosphate amidotransferase, the first enzyme in de novo purine biosynthesis (525) (Fig. 8). Ascospores from homozygous ad-7 x ad-7 crosses are white (allele Y175M256). Strains carrying allele P73Bl7l(t) are temperature sensitive.
ad-8: adenine-8
VIL. Right of ser-6 (15%) and het-8 (12%). Left of
aro-6 (8%) and cpl-1 (6 to 11%)
(437, 510, 730, PB).
Requires adenine; cannot use hypoxanthine (526). Lacks adenylosuccinate synthase (511) (Fig. 8). Fine-structure mapping and intralocus complementation (510-512). Has little hypoxanthine uptake and little hypoxanthine phosphoribosyltransferase; both these effects are partly counteracted in ad-1 ad-8 double mutants (903). Little hypoxanthine phosphoribosyltransferase is also found in mep(3) and mep(10) mutants, q.v. Used to study purine transport (787, 903, and references therein). Called complementation group E.
ad-9: adenine-9
IR. Right of met-6 (9 to 16%). Left of Tp(T54M94) and
nit-1 (3 to 15%) (466, 816).
(815)
Uses adenine or hypoxanthine (526). Controls conversion of phosphoribosylglycineamide to phosphoribosylformylglycineamide (120) (Fig. 8). Called complementation group D.
adg: adenine-arginine
See arg-11.
adh: adherent
VIIL. Linked to do (0/53), spco-4 (4%), and nic-3
(11%) (816, PB).
Abnormal morphology. Conidia not powdery; do not shake loose. Complements
spco-4.
Morphologically distinct from do and spco-4 mutants.
(816, PB).
ads: adenine sensitive
VI. Linked to col-4 (513)
Growth completely inhibited at 35°C by 10 mM adenine; high concentrations inhibitory at 25°C. Poor growth on minimal medium at 35°C as compared with that at 25°C. Inhibition not relieved by vitamins, amino acids, guanine, guanosine, or guanylic acid; no growth response to guanosine in the absence of adenine (513; T. Ishikawa, personal communication).
aga: arginase
VIIR. Between wc-1 (2%) and arg-10 (24 to 27%) (240,
697).
Presumed structural gene for arginase (240, 697) (see Fig. 10). Unable to form ornithine from arginine; arginine is thus unable to satisfy the proline requirement of pro-3 in a pro-3 aga double mutant. Prototrophic single mutants develop polyamine requirement in the presence of arginine. This is due to feedback inhibition of ornithine biosynthesis by arginine, combined with a catabolic block in ornithine formation from arginine (240). Siderophore production is severely reduced in the absence of ornithine in the triple mutant aga arg-5 ota, which has been used to study iron transport (1146, 1147) and to obtain mutants defective in siderophore uptake (G.W. Charlang and N.P. Williams, personal communication); see sit.
age-1: aging of conidia-1
IR. Symbol used for a series of many linked loci distal to nit-1, possibly
redundant
complexes. Individual loci symbolized as 1.3, 1.5, etc. Prototype age 1.5 is
right of so
(14%)
and left of aro-8 (7.6%); age 1.3 maps at same site as so,
q.v. (K.D. Munkres,
personal
communication).
Reduced conidial longevity in the light. Not expressed in the dark, or in the light with vitamin E or reduced glutathione. Deficient in an isozyme of catalase, in mitochondrial superoxide dismutase, and in other enzymes involved in destroying free radicals and peroxides. Scored by plating efficiency after incubation of mature slant cultures at 30 C, 100% relative humidity, in continuous white fluorescent light, 24 J/square meter. Also scored by a defect in conidiophorogenesis on Vogel sorbose- sucrose plates. Initial mutants selected as spontaneous variants from f1 of Oak Ridge wild types; variants with increased conidial longevity can also be selected. High spontaneous mutation rate. Longer life span correlated with slower growth. (702, 704, 705, 708; K. D. Munkres, personal communication)
age-2: aging of conidia-2
VIR. Right of ws-1 (8%). (K. D. Munkres, personal
communication)
Phenotype similar to that of age-1 (702, 704, 708).
age-3: aging of conidia-3
IR. Right of age-1 and un-18. (705, K.D. Munkres,
personal communication)
Reduced conidial longevity. Differs from age-1 and age-2 mutants in having yellow conidia and normal genesis of conidiophores (705).
al: albino
Mutants designated as albino impair carotenoid synthesis. These affect only
vegetative cells (mycelia and conidia) and are without known effect on the
perithecia or ascospores, where the pigment is melanin. The albino mutants
vary in amount and color of carotenoids. Different alleles result in conidia and
mycelia that are white, yellow, pink, purple, or white with traces of color or in
white mycelia with pigment in the peripheral conidia. See, for example,
reference 1042. Carotenoid synthesis is also affected by ylo, wc, and
age-3,
q.v., and by modifiers of intensity (982).
Albino mutants have been used to study the role of carotenoids in photoprotection (984, 1071, and references therein). Rapid development of carotenoids is induced by light; the action spectrum is described in references 250 and 1181, and mechanism of photocontrol is considered in reference 444. However, carotenoid synthesis can proceed slowly in complete darkness. Maximum carotenoid production results if incubation is at 6°C immediately after exposure to inducing light (442). Albino mutants can be scored in submerged colonies from plated ascospores by transfer of sorbose plates to 4 C under light after colonies have grown at 25°C in the dark (154, 500). An unstable constitutive variant has been described (587). Most al mutations map in a short region of IR where al-1 and al-2 were previously thought to be contiguous but are now known to be separated by other loci (797; D.D. Perkins, unpublished data).
al-1: albino-1
IR. Right of hom (<1%), arg-6 (<1 to 4%),
T(T54M94),
and al-2. Left of
lys-3 (9%). (797, 808; D.D. Perkins, unpublished data). (482)
Carotenoids abnormal. Strains carrying the various alleles differ widely in phenotype, ranging from white (e.g., 4637) and "aurescent" (pigment in peripheral conidia and conidiophores, 34508) to yellow mycelia and conidia (e.g., ALS4 and RES-25). See, for example, reference 1042. Strains carrying alleles ALS-14, RES-6, 34508, and RES-25 contain large amounts of phytoene (99 to 100% of the total neutral carotenoids), suggesting a lesion that affects phytoene dehydrogenase (398, 1039) (see Fig. 9). Strains carrying allele RWT-ylo accumulate zeta carotene and smaller amounts of neurosporene, suggesting a leaky block of the step between these intermediates (1071). It is not known whether phytoene dehydrogenase catalyzes the whole series of dehydrogenations or whether leakiness of this enzyme accounts for the different mutant phenotypes. For complementation tests, see references 500, 1039, and 1041. Fine-structure mapping (500, 1042). Translocation T(4637), inseparable from al-1, was the first albino mutation and one of the first chromosome rearrangements in Neurospora to be identified and studied (656). Allele 34508 called aur: aurescent.
al-2: albino-2
IR. Right of os-5 (<1%) and T(STL76). Left of
arg-6
(1%) and al-1 (797,
802, 808, 816, 818). Included in duplications from Tp(T54M94), confirming
location left of arg-6 (808). (482)
Carotenoids absent or abnormal, but steroids produced (398). Blocked in microsomal fraction and defective in phytoene synthetase (445), a particulate enzyme (445 and references cited therein) (Fig. 9). Tracer experiments indicate a lesion between prephytoene pyrophosphate and phytoene (572). Alleles include those resulting in white and pale rose-white, e.g., 15300 and Y254MI65 (1042), and purple, e.g., MN58a (154). For complementation, see references 500 and 1041. Fine-structure mapping (500, 1042) needs reevaluation because of new information on the location of the arg-6 marker (797).
al-3: albino-3
VR. Between his-1 and inl (1%) (1119, PB).
Carotenoids deficient (398). Reported to lack geranylgeranyl pyrophosphate synthetase activity and is blocked in soluble fraction, consistent with lesion between isopentenyl pyrophosphate and geranylgeranyl pyrophosphate (445), but can still produce farnesyl pyrophosphate (445) and steroids (398). (See Fig. 9.) This evidence contradicts in vivo labeling results that indicate a lesion between prephytoene pyrophosphate and phytoene (572). Strains carrying allele Y234M470 (al-3ros), formerly called rosy (49), become partially pigmented but are readily distinguished from the wild type. ylo-1 can be scored in combination with al-3ros (Y234M470) (PB). Strains carrying other alleles (e.g., RP100) (1119) are white with a trace of pink pigment.
FIG. 9. Biosynthetic pathway for carotenoids. It is thought that the same prenyl transferase catalyzes all the steps from dimethylallyl pyrophosphate to geranylgeranyl pyrophosphate (444; R.W. Harding, personal communication), and it has been proposed that a separate prenyl transferase converts dimethylallyl pyrophosphate to farnesyl pyrophosphate for sterol synthesis (445). The conversion of phytoene to the various carotenoid pigments involves a series of dehydrogenations, cyclizations, and other reactions. There must also be a cis/trans isomerization analogous to that found in tomato (842). The sequence of some of these steps is still uncertain; the pathway must branch, and there may be alternate routes to some of the products. See references 228, 443, 444, 842 and citations therein for proposed sequences. al-1 is probably blocked in phytoene dehydrogenase (398). It is not known whether this enzyme catalyzes the whole series of dehydrogenations. al-2 is reported blocked between geranylgeranyl pyrophosphate and phytoene (445) and between prephytoene pyrophosphate and phytoene (572). al-3 is alternately reported blocked between isopentenyl pyrophosphate and geranylgeranyl pyrophosphate (445) and between prephytoene pyrophosphate and phytoene (572), but it is not blocked in the production of farnesyl pyrophosphate or sterols (398, 445). ylo-1 is evidently blocked in a late step, probably either in the conversion of lycopene to 3,4-dehydrolycopene or in the conversion of either torulene or gamma-carotene to neurosporaxanthin (see citations in reference 398).
alc-1: allantoicase-1
II - Linked to pe (10%), probably on the opposite side of pe from
xdh-1 (24%)
(872).
Defective in purine catabolism. Unable to use allantoic acid as the sole nitrogen source. Lacks only allantoicase (872) (see Fig. 24).
alcoy
Genotype: T(IR;IIR)4637 al-1; T(IVR;VR) R2355, cot-1; T(IIIR;VI)I, ylo-1
(816).
Linkage tester containing three unlinked reciprocal translocations, each tagged with a visibly scorable marker, marking linkage groups I through VI. Linkage of a gene to al-1, cot-1, or ylo-1 in a cross to alcoy allows assignment to a linkage group by a single follow-up cross. A majority of new point mutations are linked to one of the alcoy markers (816). An improved version, alcoy;csp-2, carries the VII marker csp-2 in addition to the three original markers (811). alcoy has been used cytologically to study the synaptonemal complex and recombination nodules (396).
aln-1: allantoinase-1
VII. (872)
Defective in purine catabolism. Unable to use allantoin or any purine intermediate before it as the sole nitrogen source. Lacks allantoinase (872) (see Fig. 24).
alx-1
See ANT-1.
alx-2: alternate oxidase-2
Unmapped.
Lacks inducible cyanide-insensitive respiration. Cannot grow on antimycin A. Complements alx-1 (ANT-1) (308).
am: amination deficient
VR. Right of ure-2 (2%) and sp (4 to 8%). Left of gul-1
(<<1%) and ace-5
(<1%) (122, 570, 579, 998). (R.W. Barratt, (cited in reference 1036)
Structural gene for nicotinamide adenine dinucleotide phosphate (NADP)-glutamate dehydrogenase (336) (see Fig. 19), for which a complete 452-residue amino acid sequence has been obtained (465). Requires a source of alpha-amino nitrogen for growth, alanine being a good supplement (e.g., reference 997). Readily scorable at 25 C; leaky at 34°C (42). Leaky growth and adaptation on minimal medium are prevented by 0.02 M glycine (782, 783) or by en(am)-1, en(am)-2, or nit-2, q.v. The am mutants show abnormal regulation of reduced nicotinamide adenine dinucleotide (NADH)-glutamate dehydrogenase and are synergistic with nit-2 in this effect (226). Some am alleles (e.g., RU1) suppress the pyrimidine requirement caused by pyr-3 (CPS- ACT+) mutations (1137). Used for the first demonstration of complementation between alleles in vivo (344) (simultaneous with independent demonstration with ad-4). In vitro complementation (342). Used for studies of complementation mechanism (199, 200, 1120). Used for fine-structure mapping (337, 338). Control of intralocus recombination by rec-3 (996-998). Used to study colinearity of the gene and gene product, internal suppressors (105, 340, 465), and the action of supersuppressors (954, 955). The functional defects in several mutant enzymes with single amino acid replacements have been defined: am1 mutant enzymes fail to bind NADPH (1120); am2, am3, am19, am130, and am131 enzymes are stabilized in the inactive conformational form (30, 200, 336, 556, 1044), and all are complementable by am1; am14 is osmotically reparable and is thought to have unstable quaternary structure (340). Used in a study showing glutamine to have a role as corepressor of uricase synthesis (1118). Used to study nitrogen assimilation and metabolism (503) and nitrogen metabolite repression (186, 291). Efficient procedure for selecting new am mutants (551). Spectrum of ultraviolet irradiation (UV)- and nitrous acid-induced mutants (554). Allele am17 has a chain-terminating codon of either the amber or ochre type at residue 313 of glutamate dehydrogenase, based on amino acid replacements in revertants and by ssu-1 (956). Allele 6 is a frameshift mutation with an insertion in the Ser5 codon (985). Allele 126 is highly unstable (553). Allele 132 is a deletion (1162). The am+ gene has been cloned in Escherichia coli (J.R.S. Fincham, personal communication) and transformed back into Neurospora (J.A. Kinsey, personal communication).
amr: ammonium regulation
See nit-2.
amy-1: amylase
See sor(T9).
amy(SF26): amylase
See exo-1.
amyc: amycelial
IL. Between ad-5 and the centromere (H.B. Howe, Jr., personal
communication). (K.C. Atwood, cited in reference 789)
Conditional morphological mutant. Recessive. On sucrose media, it forms dotlike granular colonies of irregular budding vesicular elements. On permissive media, made either with acetate (plus alpha-ketoglutarate, succinate, malate, or certain amino acids) or with amino acids as carbon and nitrogen sources, it forms hyphae and macroconidia but is apparently still colonial (281; for a review, see reference 1088). Adenosine 3',5'-monophosphate induces conidiation even on sucrose (281). Photographs (774, 1088). Low oxygen consumption and depressed amino acid pools (1281). Abnormal mitochondria (773). Surface glycopeptide (281). Wall composition (207). Recovery of the antigenic arc representing the isozyme of malate dehydrogenase associated with conidiation (784). Ultrastructure (773, 1088, 1089 and references cited therein). Used extensively in balanced heterokaryons to detect lethal recessive mutations (e.g., reference 32) and to evaluate nuclear distribution (33); techniques described (34).
Last modified 4/24/96 KMC