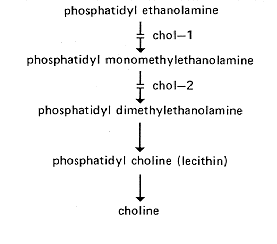
FIG. 12. Biosynthetic pathway of choline. showing sites of gene action (222, 924).
Colonial mutants (col)
Colonial-Temperature Sensitive (cot)
Crisp (cr)
Conidial Separation (csp)
Cytochrome a (cya)
Cysteine (cys)
Cytochrome (cyt)
c
Used as a symbol for het-c, cy, and col-4.
caf-1: caffeine resistant
VL. Right of T(OY321) (11%), T(AR30) (19%), and. hence, of
NO. Left of T(AR33)
(5 to 12%) and lys-1 (4 to 14%). (817, PB; K.S. Hsu, unpublished
data)
Resistant to caffeine (494). Resistance is dominant in duplications from T(AR33) (817). Scoring is clear at 25°C and poor at 34°C on slants with 2 or 2.5 mg of caffeine per ml of minimal medium without sorbose (R.L. Metzenberg, personal communication). Also readily scorable by using conidial suspensions spotted on plates containing 2 mg of caffeine per ml of minimal sorbose medium (PB).
can: canavanine resistant
See cnr.
car: carbohydrate
IVL. Linked to cys-10 (1%) (435).
Altered morphological rhythm associated with a deficiency in low-affinity glucose transport system. On glucose, the mutant car produces dense and sparse mycelia in cycles (period, ca. 50 h). Originated from a cross between pat and acu-7, but acu-7 is not necessary for the phenotype. Periodicity is affected by composition of the medium rather than time, so that the cycle is not circadian as in bd or pat mutants. On acetate, the mutant car is insensitive to the light/dark cycle and has a normal conidiation cycle with a period of about 24 h. Called LPcar: long-period carbohydrate (435). The symbol car was also used formerly for some carotenoid mutations, at least one of which is an al-2 allele (1041), and for a carbohydrate mutation (1030) strain not available for testing.
cel: chain elongation
IVR. Linked to pan-1 (1%) and cot-1 (0/17) (812).
Requires saturated fatty acids. Defect attributed to impaired chain elongation (455). Deficient in the fatty acid synthetase complex (317). increased sensitivity to oligomycin (L.R. Forman and S. Brody, cited in reference 293), Tween 20 provides a convenient supplement. Requirement is "leaky" at 21 to 22°C (317) but not at 34°C (812). Used to change fatty acid composition (113) and make the circadian clock sensitive to fatty acids and temperature (114, 649). Temperature compensation of the clock is lacking in cel mutants (650). Used to study membrane lipid-phase transitions and electrical properties (363, 364). Used to incorporate photolabile azido fatty acid probes for membrane studies (176). Called ol (oleic acid) (812) and fas (fatty acid synthesis) (317). Initial report of oleic acid utilization (812) was incorrect (455), probably because of impurities.
cell-1: cellobiase/cellulase
Unmapped. Segregates as a single gene, independent of gluc-1.
Constitutive production of both cellulose and cellobiase. Does not affect levels of aryl-p-glucosidase. Recessive to the wild type in heterokaryons (728). Isolated by using gluc-1 and selecting for high activity in destroying the beta-glucoside esculin (300). (Cellulase, cellobiase, and aryl-beta-glucosidase are normally induced simultaneously by cellobiose.
Centromeres
Three methods are available for mapping centromeres relative to flanking gene loci: (i)
tetrad analysis with ordered asci (or in unordered asci having other known centromere
markers); (ii) duplication coverage of the flanking gene in a segment carrying gene loci
known to be located in a given arm; (iii) cotranslocation of the flanking gene, together
with
other loci whose arm is known, to the arm of a known centric chromosome. (See
reference
808.) Method ii is the least laborious when appropriate rearrangements are available for
a
"left-right" test.
Linkage group arms were defined as left and right (R) (47), using mt, pe, ser-1, pdx-1, ilv-1, rib-1, and nt as reference markers.
Centromere I
Right of T(39311) and arg-3 (2%). Left of T(AR173)
and his-2 (<1%). (391,
808,1005, PB; P. St. Lawrence, cited in reference 47, 789, or 812) (Map
distance between arg-3 and his-2 varies from 3 to 18 units,
depending on rec
genes [174], so that the arg-3 and his-2 centromere distances can be
larger than
indicated.) sn and os-4 also lie between these translocations and
are, therefore,
the gene markers closest to the centromere.
Centromere II
Probably between bal and arg-5, from ordered asci (428; L.
Garnjobst,
personal communication). Right of T(AR179) and left of
T(ALS176) (808,
PB); bal also lies between these translocations.
Centromere III
Left of sc and near thi-4 by ordered asci (47). Order is uncertain
relative to
acr-2, q.v.
Centromere IV
Left of psi (D.R. Stadler, A.M. Towe, and M. Loo, cited in reference 619) and
T(ALS159) (808). Right of cut and fi by inference from
recombination
distances, but no direct evidence.
Centromere V
Right of T(AR33) and, hence, of caf-1. Right of
T(AR30) (817). Near and
perhaps right of lys-1 by ordered asci (47). Left of per-1 (18%) by
ordered asci
(489).
Centromere VI
Right of ad-1 (1 to 2%) and ylo-1 (1 to 6%). Left of
T(AR209), rib-1 (1 to
4%), and pan-2 (2%) (138, 808, 1012). (glp-4 is also between
ad-1 and rib-1
[1102].)
Centromere VII
Right of T(T54M50) and, hence, of csp-2 and thi-3 (808).
Left of met-7 (<1%,
one critical ascus) (M.E. Case, personal communication). Near sfo (<1%)
and
qa (no recombination) by ordered asci (318; M.E. Case, personal
communication).
cfs(OY305): caffeine sensitive
IR. Between mating type and al-2 (1172).
Growth inhibited by caffeine (0.2 mg/ml) on minimal medium. Growth stimulated by adenine. Not sensitive to caffeine in the presence of adenine. Slow growth on minimal medium. Morphologically abnormal. Probably UV sensitive by spot testing. Not tested for allelism with ad-3A, ad-3B, or ad-9 (1172).
cfs(OY306): caffeine sensitive
IR. Near al-2, probably to the right (1172).
Growth inhibited by caffeine (0.2 mg/ml) and by adenine. Slow growth on
minimal medium. Not stimulated by complete medium. Morphologically
abnormal. Complements cfs(OY305) and cfs(OY307).
(1172)
cfs(OY307): caffeine sensitive
IR. Near cfs-1 between mating type and al-2 (1172).
Growth inhibited by caffeine (0.2 mg/ml) and by adenine. Slow growth on minimal medium. Not stimulated by complete medium. Morphologically abnormal. Complements cfs(OY305) and cfs(OY306). (1172)
chol-1: choline-1
IVR. Linked to ad-6 (1%). probably to the right (633). (482)
Requires choline (470). Also uses mono- or dimethylaminoethanol (468) (Fig. 12). Deficient in S-adenosylmethionine:phosphatidylethanolamine-N-methyl transferase (222, 924). Abnormal colonial morphology on limiting choline (222). Colonies from single conidia on minimal agar medium resemble inhibited A/a duplications, with swollen hyphae and darkening in the presence of phenylalanine plus tyrosine (D.D. Perkins, unpublished data). Abnormal phospholipid composition on limited concentrations of supplement (501). Best scored late on minimal medium. Grows slightly and then stops (D.D. Perkins, unpublished data). Used to study inhibition of cytochrome-mediated respiration and of conidiation when lecithin is depleted by choline starvation (534, 535). Initial allele called 34486.
chol-2: choline-2
VIL. Left of nit-6 (6 to 8%) (812, PB).
Requires choline (471). Also uses di- but not monomethylaminoethanol (468) (Fig. 12). Deficient in S-adenosylmethionine:phosphatidyl- monomethylethanolamine methyltransferase (222, 923, 924). Strains carrying the only allele, 47904t, are leaky on minimal medium at 22°C but not at 34°C (501). Phospholipid composition is abnormal on limiting choline (501). Growth is colonial on limiting supplement at 34°C and on minimal medium at 25°C.

FIG. 12. Biosynthetic pathway of choline. showing sites of gene action (222,
924).
chr:chrono
VI. Between chol-2 (10%) and pan-2 (376).
Altered period of circadian conidiation rhythm. The one allele known is incompletely dominant, specifying a 23.5-h period at 25°C in a csp+ genetic background. Temperature compensation good above 30°C (375, 377).
cl: clock
VR. Right of pk (<1 %, 2%) (296, 1007).
Spreading flat colonies, forming dense bands. Noncircadian periodicity. Mycelium becomes increasingly dense until growth ceases in all but a few hyphae, which reinitiate the cycle. Grows 1 cm per day. Band size and period modifiable. Photographs (296, 1046). Asci from cl x wild type crosses are normal. Homozygous cl x cl crosses (with backcrossed derivatives) give flaccid asci with unordered ascospores similar to those from pk x pk crosses. cl x pk crosses do also, suggesting allelism (1007). pk (bis) mutants sometimes form growth bands (296). However, substantial crossing-over frequencies and the recovery of pk cl double mutants (296) indicate that pk and cl are at separate loci. Increased activity of L-glutamine:D-fructose-6-phosphate amidotransferase was found in crude extracts of cl and five other morphological mutants (899).
cni-1: cyanide insensitive-1
Unmapped chromosomal gene. Not in V (309).
No cyanide-sensitive or antimycin A-sensitive respiration in the first 24 h of growth. Also, initially insensitive to salicyl hydroxamic acid. But salicyl hydroxamic acid and cyanide together inhibit (309). Cyanide-insensitive respiration and the cytochrome c level decrease markedly in the late log phase and stationary phase, whereas the cytochrome aa3 level increases rapidly (559). As shown by electron microscopy, mitochondria are defective in the early log phase, but later resemble the wild type (558). Behaves as a cold-sensitive ribosome assembly mutant. Small subunits are not assembled at low temperatures; the respiratory differences between early and late growth noted above were found at 30°C, and normal aa3 production occurred throughout growth at 37°C (548). Electron spin resonance data (308). Selected by failure to reduce tetrazolium in overlay after inositol-less death enrichment (310).
cnr: canavanine resistant
IR. Linked to hom (1%) and nic-1 (11%), probably between them
(812).
(1061)
Resistance to canavanine (480, 1061, 1062) is due to a constitutive enzyme that cleaves L-canavanine to hydroxyguanidine plus a compound which can be converted to L-homoserine (617). Resistance is altered secondarily by modifiers that affect rate of uptake (617). Many laboratory strains are resistant, but a few are sensitive (790). Best scored on 0.2 mg of L-canavanine H2SO4 per ml (autoclaved in the medium) at 2 days, 34°C (812; D.D. Perkins, unpublished data). Sensitive strain used to select resistant mutants defective in basic amino acid transport (889, 913, 1152); see pmb. Called can.
cog: recognition
IR. Between his-3 (1 to 3%) and ad-3A (2 to 7%) (27,
171).
The postulated site for initiating local recombination in meiosis. Affects recombination in adjoining regions, in the absence of rec-2+. Presence of cog+ then increases recombination within his-3 (27) and crossing over between his-3 and ad-3 (171). The allele for high recombination is completely dominant, and its effect is specifically on the his-3 alleles in chromatids that contain cog+, as evidenced by crosses heterozygous for reciprocal translocation TM429, which has a breakpoint between sites within the his-3 locus (171). See rec and rec-2; see Fig. 22.
coil
IVR. Between arg-2 (2%) and leu-2 (12%) (74).
Hyphae grow in clockwise coils on the surface of agar medium. Scorable in young cultures microscopically under low magnification (74).
col: colonial
Name used primarily for mutants having restricted mycelial growth that is
self-limiting on agar medium. Usually, radial growth of each colony does not
exceed a few millimeters. Colonial mutants vary widely in texture, density,
conidiation, pigment, and fertility. Mutations of the series col-5 to
col-17,
described by Garnjobst and Tatum (382), were in some instances assigned new
locus names without having been tested for allelism with already named
morphological mutations having similar map locations. Some were shown
subsequently to be recurrences (e.g., col-7 of rg-1; col-14
of sc); others may be
recurrences but have never been tested (possible examples: col-5 and
col-8
with col-1; col-13 and col-15 with vel. Two
colonial mutants with
nongerminating ascospores are symbolized le-1 and le-2. For
reviews covering
morphological mutants and morphogenesis, see references 112, 642, 675, 942,
946, 1088. Growth rates and hyphal diameters of numerous colonial mutants
are given in reference 197.
col-1: colonial-1
IVR. Linked to pan-1 (0/47 asci) and cot-1 (3%) (46,
374).
Colonial morphology, with no macroconidiation. Growth cyclic at moderate temperatures; steady at 38°C. Double mutant col-1 cot-1 grows better than col-1 at 24°C and better than cot-1 at 38°C (374). The double mutant pe;col-1 forms microconidia (46) and was used in early mutation studies, but col-4;pe fl was found to be better (386). It was found (416) that pe;col-1 strains form microconidia at 25°C but form macroconidia at 35°C. Cell wall analysis; photograph (278). col-5 and col-8 not tested for allelism.
col-2: colonial-2
VII. Linked to met-7 and met-9 (1%), probably to the left
(812, 816). Right
of T(T54M50) (D.D. Perkins, unpublished data).
Colonial morphology (46). Photographs (112, 9469 948). Altered structure of NADP-specific glucose-6-phosphate dehydrogenase (116, 949). (bal and fr mutants are also deficient in glucose-6-phosphate dehydrogenase.) Reduced NADPH level (110). Reduced linolenic level (115). Accumulates much neutral lipid (765). Pyridine nucleotide levels (111). Suppressor su(col-2) increases the linear growth rate and influences the electrofocusing pattern of col-2 glucose-6-phosphate dehydrogenase (948). Homozygous col-2 x col-2 crosses fail to mature. Immature asci are frequently nonlinear and occasionally show dichotomization (1007). Hyphae swell with age to diameters of 20 um (197).
col-3: colonial-3
VII. Linked to met-7 (0/93) and wc-1 (1%) (812).
Complements col-2 in
heterokaryons (K. Wilson, cited in reference 46). Right of T(T54M50) (D.D.
Perkins, unpublished data).
Colonial morphology (46). Altered 6-phosphogluconate dehydrogenase (947, 949). (col-10 also affects 6-phosphogluconate dehydrogenase.) Reduced NADPH level (110). Reduced linolenic acid level (115).
col-4: colonial-4
IVR. Between met-1 (4%) and arg-2 (<1 to 2%) (692,
876, 991). (695)
Spreading colonial morphology, forming dense balls of conidia high in slants (47). Probably dominant in heterozygous duplications from T(S1229) (E.G. Barry, personal communication). Cell wall-autolyzing enzyme (631). Reduced amount of cell wall peptides (1165). Used in combination with pe fl to produce microconidiating colonial growth suitable for reversion experiments (386). Called spco-1 (382); called c (386).
col-5: colonial-5
IVR. Linked to cot-1 (1%), probably to the right (819). (382)
Dense, nonconidiating, poorly pigmenting, slow spreading, colonial morphology (382, 819). Cell wall analysis and photograph (278). Reverts readily (382). Not tested for allelism with col-7, which it resembles and which maps in same region. Called col(B28) (819).
col-6: colonial-6
IV. Linked to the centromere (0/28 asci) and pan-1 (21%) (382).
Colonial morphology. Slow ascospore germination (382).
col-7: colonial-7
Allelic with rg-1, q.v. (675).
col-8: colonial-8
IVR. Linked to pan-1 (4 to 13%) (382).
Colonial morphology, with fluffs of hyphae top of slants. col-8 x col-5 crosses not fertile. Not tested for allelism with col-1. (382) Reduced amount of peptides in cell wall (1165).
col-9: colonial-9
VR. Between inl (16%) and asn (5%) (698).
Small, slow-growing colony. Reverts readily (382).
col-10: colonial-10
IIL. Linked to cys-3 (14%), near or at pi. One wild type in 81
progeny from
a col-10 x pi cross may have been a revertant (382, 816).
Slow growing; dense, compact morphology without conidia (382). Altered 6-phosphogluconate dehydrogenase (947, 949). (col-3 also affects 6-phosphogluconate dehydrogenase.) col-10 (R2438) x pi (B101) crosses resemble R2438 x R2438 and B101 x B101 crosses in producing abnormal asci with flaccid walls and unordered ascospores, suggesting allelism (1007). However, R2438 and B101 mutants are distinctly different in morphology. Mutant B101 has not been tested for 6-phosphogluconate dehydrogenase. As a marker, pi (B101) is preferable to R2438 because of growth rate, stability, and ease of handling; ro-7 in the same region is preferable to both (PB).
col-12: colonial-12
I. Linked to mt (17 to 22%) (382).
Colonial morphology. Slow growth from ascospores (382).
col-13: colonial-13
IIIR. Putative vel allele. Linked to tyr-1 (4%) and
col-16 (0/181) (382).
No direct intercross with vel.
col-14: colonial-14
Allelic with sc, q.v. (PB).
col-15: colonial-15
IIIR. Putative vel allele (0/26) (PB). Linked to tyr-1 (5%) and
col-13 (0/181)
(382).
Resembles col-13 and vel. Occasionally forms puffs of aerial conidia at tops of slants; these are not due to reversion (PB). Reported to complement col-13 in heterokaryons (382), but this may have been due to misinterpretation of erratic conidiation.
col-16: colonial-16
IIIR. Linked to leu-1 (1%) and pro-1 (10%) (382; PB).
Colonial morphology (382). Forms balls of powdery conidia at top of slants of glycerol complete medium. A good marker, preferable to com; strains carrying the latter grow more slowly and do not conidiate (PB). Complements mo-4 and spco-15 (382).
col-17: colonial-17
VII. Linked to nt (14%) and spco-5 (6%) (382).
Colonies grow very slowly (382).
col-le: colonial, lethal ascospore
See le-1.
com: compact
IIIR. Between ace-2 (5%) and ad-4 (<1 to 5%) (578,
814).
Forms small, slow-growing colonies (278, 789). Grows better on complete medium than on minimal medium (PB). Cell wall analysis; photograph (278). Called B54.
con-1: recombination control-1
Element postulated to lie near nit-2 (IL) and also proximal to his-1
(VR),
interacting specifically with the rec-1 gene product to regulate recombination.
No genetic variants are known. (170)
con-2: recombination control-2
Element postulated to lie between his-3 (IR) and ad-3, in or left of
the interval
arg-3 to sn (I) and also between pyr-3 and
his-5 (IVR), interacting specifically
with the rec-2 gene product to regulate recombination. No genetic variants
are
known. (170)
con-3: recombination control-3
Element postulated to lie proximal to am-1 (VR) and also between
sn and
his-2 (IR), interacting specifically with the rec-3 gene product to
regulate
recombination. No genetic variants are known. (170)
con: (conidiation)
See cr-1.
cot-1: colonial temperature sensitive-1
IVR. Between pan-1 (2%) and his-4 (1 to 6%) (692, 812,
816).
Extremely colonial at 34°C, but completely normal growth, morphology, and fertility at 25°C and below. Linear growth is maximum at 24°C (374). Becomes colonial at 32°C; colonies from ascospores or conidia are viable and continue to grow slowly with dense branching, but do not conidiate. They quickly resume normal growth when shifted to a permissive temperature (692, 1068). Recessive in duplications (808); apparent dominance in heterokaryons (374) may have resulted from a shift in nuclear ratios. Used in studies of septation and branching (202), growth-inhibiting mucopolysaccharide (878, 879), and sulfate transport (641). Cell wall analysis (374). Growth is stimulated by lysine or arginine (0.1 mM) on glucose media at high temperatures (615).
Because of high viability and tightly restricted growth at restrictive temperatures and normality at 25°C, cot-1 mutants have valuable technical applications. For example, crosses homozygous for cot-1 have been used in combination with sorbose for experiments with rec genes, where high-density ascospore platings are required for precise quantitative analysis of intralocus recombination (e.g., references 165, 997, and 1070). In another application, when shifted up after initial growth at the permissive low temperature, cot-1 hyphae assume a "bottle brush" appearance with small side branches (692). This has been used to select uvs mutants by subsurface survival on UV-irradiated plates containing p-aminobenzoic acid (938; D.E.A. Catcheside, personal communication). cot-1 conidia or ascospores from cot-1 x cot-1 crosses are used for replication in a protocol involving transfer by filter paper (615). For suppressors of cot-1, see gul.
cot-2: colonial temperature sensitive-2
VR. Right of pk (8%) and ser-2 (5%). Left of ad-7 (4%)
(156, 818). (698)
Recombines with inv (5%) (315).
Small colonies at 34°C but fully viable. Growth and morphology nearly normal at 25°C but not completely so (382). Makes altered invertase; it is not clear whether cot-2 is the structural gene for a second subunit or whether cot-2 affects structure indirectly, e.g., by altering the carbohydrate moiety (315). See inv. Reduced amount of cell wall peptides (1165). Ascospores are normal in heterozygous crosses, but are round in homozygous cot-2 x cot-2 crosses (59). Some cot-2 strains carry mei-3, which was found in the original cot-2 strain (757); however, most strains used by Eggerding et al. (315) are free of it, and mei-3 cannot be responsible for the effects on invertase (D. Newmeyer, unpublished data).
cot-3: colonial temperature sensitive-3
IV. Left of pan-1 (16 to 25%). Linked to arg-2, probably to the
right. (382,
PB)
Small colonies at 34°C, but fully viable. Growth and morphology normal at 25 C (382).
cot-4: colonial temperature sensitive-4
VR. Right of ilv-1 (8%) and rol-3 (5%). Left of inl
(10%) (698). Not allelic
with sp (11%) (PB).
Small colonies at 34°C, spreading at 25°C (382). Morphology at 25°C resembles that of the mutant sp, with late-forming blooms of conidia on aerial hyphae, but sp is not heat sensitive. Good female fertility, but no perithecia in homozygous cot-4 crosses (PB).
cot-5: colonial temperature sensitive-5
IIL. Right of T(P2869) and T(B18). Probably left of
pyr-4 (0/39). Linked to
fs-1 (29%) (PB). (382)
Little or no growth at 34°C; colonial at 30°C. Morphology still not normal at 25°C, a temperature at which older colonies form short aerial hyphae (382). Female sterile. Morphology distinct from that of the mutant fs-1 (PB).
cpc-1: crosspathway control
VI. Right of ylo-1 (1 to 3%) (238).
Affects simultaneously both ability to derepress and basal levels of enzymes in arginine and other amino acid biosynthetic pathways. cpc-1 mutations interfere with cross-pathway control of amino acid biosynthetic enzymes. Sensitive to 3-amino-1,2,4-triazole. Isolated as arginine auxotrophs by selection in the mutant arg-12s, q.v. The cpc-1;arg-12+ single mutant is prototrophic. Delayed growth after ascospore germination; the delay is not alleviated by arginine or precursors. Scorable by delayed growth. (61, 238) Exemplified by alleles CD-15 and CD-55 in reference 238 and j-2, j-5, and j-9 in reference 61.
cpl-1: chloramphenicol sensitive-1
VIL. Between ad-8 (6 to 11%) and lys-5 (6%) (180, PB).
Sensitive to chloramphenicol (< 0.5 mg/ml added to autoclaved medium) and to antimycin A (1 µg/ml). (The wild type is resistant to 4 mg of chloramphenicol per ml.) Protein synthesis is not grossly altered. Cyanide-insensitive and azide-insensitive respiratory systems are still present. Cytochrome spectrum normal on minimal medium. Obtained by inositol death enrichment and replica-plating, using a strain of genotype inl;trp-3;sn cr-1 (180, 182). Scorable on 0.5 mg of chloramphenicol per ml autoclaved in medium (PB).
cpt: carpet
IIR. Right of arg-5 (3%). Left of T(NM177) and pe
(6%)
(812, 808).
Flat, slow-growing mycelium with no macroconidia. Produces microconidia (812; S. R. Gross, personal communication), but much less abundantly than do the double mutants fl;dn and pe fl (PB). Homozygous fertile.
cr-1: crisp-1
IR. Right of ace-7 (1 to 3%) and nic-2 (4 to 7%). Left of
cys-9 (3%) and un-1
(5%) (721, 816). Included in duplications from T(4540), which do not include
cr-2 or cr-3 (PB). (610)
Rapid conidiation close to surface of agar. Produces very short conidiophores, bearing conidia in tight clusters (610, 611). Photographs (533, 634). Recessive. Deficient in adenylate cyclase (1066); has little or no endogenous adenosine 3',5'-phosphate (1065, 779). Abnormal morphology partially corrected by exogenous adenosine 3',5'-phosphate (891, 892, 1065, 1066). Guanosine 3',5'-phosphate also stimulates mycelial elongation (892). Cyclic nucteotide levels differ in mycelia and conidia (891, 892). NAD(P) glycohydrolase is overproduced and excreted; this is normalized by adenosine 3',5'-phosphate (533). Induction and localization of p-glucosidase is altered; induction is normalized by adenosine 3',5'-phosphate (906). Inability to use glycerol and certain other carbon sources is also overcome by adenosine 3',5'phosphate (598, 1067). Phosphodiesterase inhibitors do not counteract the morphological effect of cr-1 (892). Increased lactate dehydrogenase activity (92). Used to determine what functions are controlled by adenosine 3',5'-phosphate (779). Used to study adenosine 3',5'-phosphate binding protein (1082).
Strains carrying the various alleles vary in growth habit (B123 strains are flat, restricted; allele L strains are spreading, but morphology may vary on different media). Modifier mutations which alter morphology and the ability of cr-1 to use glycerol occur frequently (383, 905). Crosses homozygous for allele B123 exude intact linear asci (634). Double mutants sn cr and cr rg form small conidiating colonies suitable for replica plating with velvet (182, 634, 796, 932, 1020). The triple mutant sn cr;csp-2 can be overlayered (744; photograph 747). The single mutant (B123) can be replicated by using a needle replicator (634). Scorability and viability are good. Excellent as a marker. Carotenoids formed normally. cr-1 ascospores may require longer to mature than cr+ ascospores. Allele CE4-11-67 called con (716, 717).
cr-2: crisp-2
IR. Right of cr-3 (11%) and T(NM103); hence, right of
thi-1. Left of al-2
(18%) (383, PB).
Conidiation delayed. Fine, pale-pigmented conidia produced in clumps over the agar surface (383). Recessive. Reduced amount of cell wall peptides (1165). Overproduces and excretes NAD(P) glycohydrolase, but this is not cured by exogenous adenosine 3',5'-phosphate (533).
cr-3: crisp-3
IR. Right of cr-1 (13%) and of T(4540); hence, right of
cys-9 and un-1. Left
of cr-2 (11%) (383, PB).
Delayed conidiation; ultimately producing fine, pale conidia uniformly over the agar surface (383). Recessive. Reduced amount of cell wall peptides (1165). Overproduces and excretes NAD(P) glycohydrolase; this is not cured by exogenous adenosine 3',5'-phosphate (533).
crib-1: cold-sensitive ribosome biosynthesis
IV. Linked to met-1 (6%) (927).
Defective ribosome biosynthesis below 20°C; attributed to a defect in ribosomal ribonucleic acid (rRNA) processing (897). Grows at 6 the wild-type rate at 10°C and 79% at 25°C. 37S cytosolic ribosomal subunits are underaccumulated, and relatively little stable 17S rRNA is produced at low temperatures. Not a conditional lethal mutation (896, 927). Conditionally defective in expression of S-adenosylmethionine synthetase activity (900).
crib(PJ31562): cold-sensitive ribosome biosynthesis
IVR. Near crib-1 (3%) (895).
Defective in biosynthesis of cytosolic ribosomes at 10°C, but normal at 25°C. Grows at 16% of the wild-type rate at 10°C and 90% at 25°C. Underaccumulates 17S rRNA and, hence, 37S ribosomal subunits. Partial complementation in forced heterokaryons with crib-1 (895). Called PJ31562.
csh: cushion
IR. Between thi-1 (12 to 20%) and ad-9 (5%) (816). (P. St.
Lawrence, cited
in references 47, 789 or 812)
Restricted colonial growth (812).
csp-1: conidial separation-1
IL. Between arg-3 (1%) and the T(39311) right breakpoint
(972, PB).
Conidia fail to separate and become airborne. Photograph (972). Recessive. Cultures on agar readily scored by the "tap test." In water, conidia are freed at 1/10 the wild-type concentration (972). Used in connection with bd for study of circadian rhythms (e.g., reference 114). Useful in student laboratories to avoid contamination (966). Carotenoids tend to be yellowish in young cultures (PB).
csp-2: conidial separation-2
VIIL. Linked to thi-3 (<1%), probably to the right. Left of
T(T54M40) (972,
PB).
Conidia fail to separate and become airborne. Cultures on agar readily scored by the tap test. Resembles csp-1. Conidia are freed in water suspension long after induction of aerial growth and at only 1/100 the concentration of the wild type. A csp-1;csp-2 double mutant releases no detectable free conidia under the same conditions (972). Most csp-2 alleles complement csp-1 in forced heterokaryons to form the wild-type number of free conidia (972), but csp-2 (UCLA102) does not (969). Conidiating colonies of the csp-2;sn cr-1 strain on replica plates can be overlayered without the conidia being spread (744); photograph (747).
cum: cumulus
IIIL. Left of r(Sk-2) (4%), acr-7 (5 to 18%), and acr-2
(18%). No
recombination with Sk-2K. (PB; B.C. Turner, personal
communication)
Initially colonial; then spreads and sends up blooms of aerial hyphae which conidiate profusely at the shallow ends of agar slants. Good female fertility. Similar in morphology to the mutants sn and sp and to the mutant cot-4 at 25 C (PB).
cut: cut
IVL. Between cys-10 (28 to 37%) and fi (4 to 10%)
(802; D.D. Perkins,
unpublished data). Linkage to IR, shown in original cut strain HK53, was due
to an unrecognized I;IV translocation (808). When an allelic cut point
mutation (LLMI) became available, it was mapped in IV rather than in I and
segregated independently of I markers (802).
Sensitive to high osmotic pressure. Phenotype similar to that of os mutants. Scorable either by morphology or by failure to grow on agar containing 4% NaCl. Morphology approaches normal at high humidity (573). Allele HK53 is inseparable from T(I;IVL)HK53 (808).
cwl: cross wall
II. Linked to arg-5 (3%) (PB).
Hyphal septa are largely absent. Hyphae tend to bleed, forming an exudate on the agar surface and in lens-shaped pockets beneath. Slow growing, aconidial. Subject to alteration by modifiers that restore septa and increase growth rate, but original mutant gene can be extracted by crossing. Recessive. Stocks conveniently kept as heterokaryons (382; A. Hammill, via FGSC; PB). Called mo(R2441).
cy: curly
IL. Linked to arg-1 (0/34) and ad-5 (1/54), probably to the left
(PB). (689)
Curly hyphae grow at wild-type rate (689). Gross morphology is indistinguishable from that of the wild type. Scorable by examining young hyphae on agar or glass walls of culture tubes before conidiation. Symbol changed from c to avoid confusion with het-c.
cya-1: cytochrome a-1
IL. Linked mating type (6%), indicated to the left (87).
Deficient in cytochrome aa3. Cannot reduce tetrazolium. Very slow growth. Female sterile (87). Possibly not really a cya mutant (H. Bertrand, personal communication).
cya-2: cytochrome a-2
VR. Linked to al-3 (3%), indicated to the right (87).
Deficient in cytochrome aa3. Cannot reduce tetrazolium. Slow growth. Female sterile (87).
cya-3: cytochrome a-3
VIL. Between chol-2 (10%) and cyt-2 (10%) (87).
Deficient in cytochrome aa3. Cannot reduce tetrazolium. Slow growth (87). aa3 deficiency suppressed by antimycin A (84). Spectrum (84).
cya-4: cytochrome a-4
IIL. Linked near thr-3 (87).
Deficient in cytochrome aa3. Cannot reduce tetrazolium. Slow growth. Spectrum (84, 87). Cytochrome oxidase subunits 5 and 6 are deficient or lacking (90).
cya-5: cytochrome a-5
IVR. Right of pan-1 (2%) (739).
Deficient in cytochrome aa3. Slow growth. Subunit 1 polypeptide of cytochrome c oxidase absent by immunological criteria. Poor recovery (10%) from crosses. Selected as tetrazolium nonreducer. (90, 739, 740) Called cya-U-34.
cya-6: cytochrome a-6
IVR. Right of pan-1 (2%) (739).
Deficient in cytochrome aa3. Alleles 2 and 35 are heat sensitive. Selected at 41°C by slow growth on salicylhydroxamic acid and resistance to tetrazolium. At least five subunits of cytochrome c oxidase are present at 41°C by immunological criteria, but are not associated. Complements cya-5. (739) Alleles called cya-6-2 and cya-6-35.
cya-7: cytochrome a-7
III. Linked to ad-4 (25%) (739).
Deficient in cytochrome aa3. Allele cya-7-13 is heat sensitive. Selected at 41 C by slow growth on salicylhydroxamic acid and resistance to tetrazolium. At least five subunits of cytochrome c oxidase are present at 41°C by immunological criteria, but are not associated. (739) Called cya-7-13.
cyb-1: cytochrome b-1
VR. Between al-3 (24%) and his-6 (10%) (87).
Deficient in cytochrome b. Cannot reduce tetrazolium. Slow growth. Spectrum (84, 87). Suppresses the aa3 deficiency of the mutant cyt-2 and of mitochondrial mutant [mi-3] (84).
cyb-2: cytochrome b-2
Unmapped. Report of VI linkage (87) may be incorrect (H. Bertrand,
personal communication).
Deficient in cytochrome b; erratic deficiency of aa3 (H. Bertrand, personal communication). Cannot reduce tetrazolium. Very slow growth. Reduced female fertility. Spectrum. (84, 87)
cyb-3: cytochrome b-3
IIL. Left of ro-3 (9%) (PB).
Deficient in cytochrome b. Cannot reduce tetrazolium. Slow growth. Heat sensitive: mutant phenotype at 38 to 39°C; nearly normal at 25 and 34°C (1133). Grows slowly from ascospores at 34°C.
cyh-1: cycloheximide resistant-1
IR. Right of nit-1 (6%). Left of T(STL76) and al-2 (8 to
13%) (496, 797, 808).
Resistant to cycloheximide (496, 748). Resistance is recessive in duplications (1090). Dominance reported in forced heterokaryons (496, 748) may have been due to skewed nuclear ratios (1090). Protein synthesis on ribosomes of the mutant cyh-1 proceeds in the presence of cycloheximide in a cell-free system (834). Readily scored on slants with 10 µg of cycloheximide per ml autoclaved in the medium. Excellent as a marker and valuable for selecting somatic recombinants or deletions in heterozygous duplications (748, 1091). Used to show that the cycloheximide-induced phase shift of the circadian clock involves protein synthesis (738). Called act-1: actidione resistant-1.
cyh-2: cycloheximide resistant-2
VR. Right of lys-2 (<1%). Left of leu-5 (<1 to 2%) and
sp (2 to 9%) (496,
818, PB).
Resistant to cycloheximide (496, 748). Protein synthesis on mutant ribosomes proceeds in the presence of cycloheximide in a cell-free system (834). Excellent marker. Readily scored on slants with 10 µg of cycloheximide per ml autoclaved in the medium or with 1 µg added after autoclaving. Resistance in heterokaryons has been reported to be dominant (496, 626) or recessive (939); it may depend on nuclear ratios or media. Used in mutagenicity test systems (626). Used to show that the cycloheximide-induced phase shift of the circadian clock involves protein synthesis (738). Double mutant cyh-1;cyh-2 grows slowly and is much more insensitive to cycloheximide than either single mutant (496).
cyh-3: cycloheximide resistant-3
Unmapped. Unlinked to cyh-2. Stated to be distinct from cyh-1
(1108).
Resistant to 100 µg of cycloheximide per ml. Double mutant cyh-2;cyh-3 is morphologically abnormal, resistant to more than 2,400 µg/ml (1108). The one known allele, CH96, was first called act-5 (1107) and then act-3 (1108).
cys: cysteine
Cysteine auxotrophs are characterized as being unable to use inorganic sulfate
but able to grow on either cysteine or methionine (721). Some cys mutants
can
use sulfite or thiosulfate. The sulfur permease mutants cys-13 and
cys-14 are
exceptions, having no demonstrable requirement. Cysteine mutant strains tend
to accumulate secondary mutations in the pathway, suggesting that some
double-mutant combinations have a selective advantage over single-mutant
strains (721). Unambiguous definition of loci based on map location is thus
important, and care must be taken that derived stocks carry the original cys
mutation. Cysteine and methionine loci provide several examples of closely
linked pairs of genes: cys-1 cys-2, cys-5 cys-11, and met-7 met-9.
With at least
two cys mutants (cys-3, cys-5), and possibly others, ascospore
maturation and
recovery of cys progeny requires that crossing medium be supplemented, even
when the protoperithecial parent is cys+. Cysteine mutants grown on limiting
supplement show a shortened period of circadian conidiation rhythm (329,
333). Partial suppressors of leu-4 which originate as double-mutant
microcolonies on minimal medium are leaky cys mutants of various types
(425); see leu.
cys-1: cysteine-1
VIL. Between cys-2 (1 to 3%) and ylo-1 (8%) (721,1012).
(980)
Uses sulfite, thiosulfate, cysteine, or methionine. Original isolate (allele 84605) also had a partial requirement for tyrosine and showed high tyrosinase activity at 25°C but not 35°C (479, 721, 980). These properties reverted, however, whereas the cysteine requirement is stable (479). Used in studies of intra- and interlocus recombination (721, 1015, 1016, 1024-1026).
cys-2: cysteine-2
VIL. Between un-4 (4%) and cys-1 (1 to 3%). Very close to or
contiguous
with cys-1, but is probably a separate locus (721, 1012). (M. Fling, cited
by T.
H. Pittenger, Genetics 39:326-342, 1954)
Uses cysteine or methionine. Strains carrying these alleles are heterogeneous in response to thiosulfate, but do not use sulfite (721). Lacks sulfite reductase, as do the cys-4 and cys-10 mutants (596). No interallelic complementation. Used in studies of intra- and interlocus recombination (see cys-1).
cys-3: cysteine-3
IIL. Right of pi (4%). Left of T(AR18) and pyr-4 (18 to
21%) (721, 808, 816).
Uses sulfite, cysteine, or methionine; little or no response to thiosulfate (640, 721). Regulator of genes of sulfur uptake and metabolism (e.g., sulfate permease, aryl sulfatase, choline sulfatase) (284, 640, 667). Grows well on methionine. Resistant to chromate (640). Used extensively for studying regulation; for a review, see reference 642. cys-3 ascospores darken slowly or not at all, even when a cys-3 strain is the fertilizing parent and when a strain carrying heat-sensitive allele NM27t is crossed at 25°C, the permissive temperature for growth. Adding methionine to crossing medium promotes darkening but fails to give good recovery of cys-3 progeny. Recovery of a few percent cys-3 progeny is possible in well-aged crosses. (721, PB). cys-3 can be used effectively as an autonomous ascospore color mutant for demonstrating segregation patterns in asci (811; see reference 858 for photograph; see Ascospore color mutants).
cys-4: cysteine-4
IVR. Right of mat (10%) and T(NM152). Left of uvs-2
(5%) (721, 808, 1023).
(815)
Uses cysteine; slight response to thiosulfate (721). Poor growth on methionine. Lacks sulfite reductase, as do cys-2 and cys-10 mutants (596).
cys-5: cysteine-5
IL. Between leu-4 and ser-3 (0.1%) (816, 1125). (N.H.
Horowitz, cited in
references 721 and 815) Probably a locus distinct from cys-11, with the
order
leu-3 cys-5 (<1%) cys-11 mt in a cross showing no negative
interference (723).
Uses sulfite, thiosulfate, cysteine, or methionine (721). Lacks 3'-phosphoadenosine-5'-phosphosulfate reductase (F.-J. Leinweber, cited in references 721 and 723). Enzymatically distinct from cys-11 (adenosine 5'-triphosphate sulfurylase), which it complements in heterokaryons (721, 723; F.-J. Leinweber, cited in references 721 and 723). Leaky, but not so as to interfere with scoring. Ascospores may be oozed from perithecial beaks rather than shot. For good recovery of cys-5 progeny, crossing media should be supplemented even when the protoperithecial parent is cys+; otherwise cys-5 ascospores may fail to blacken. cys(NM86) and cys(85518), initially listed as cys-5 alleles, are now designated cys-11.
cys-6, -7, -8: cysteine-6,-7, -8
Lost. Identity or nonidentity with other loci was never established (721, 823).
cys-9: cysteine-9
IR. Between cr-1 (3%) and thi-1 (13%) (721).
Uses sulfite, thiosulfate, cysteine, or methionine. Somewhat leaky. (721)
cys-10: cysteine-10
IVL. Left of acon-3 (1 to 6%), ace-4 (19 to 33%), and
cut (28 to 37%) (578,
PB). (721)
Uses cysteine, cystathionine, homocysteine, or methionine, with a slight response to thiosulfate (469, 596, 721); however, E. Kafer (personal communication) found good growth on thiosulfate. Growth is better on casein hydrolysate than on methionine (D.D. Perkins, unpublished data). cys-10 chol-1 double mutants grow better on methionine alone than on methionine plus choline (721). Lacks sulfite reductase, as do cys-2 and cys-4 mutants (596). Formerly called met-4; see reference 721.
cys-11: cysteine-11
IL. Linked to cys-5 (<1%), between leu-3 (8%) and mating type
(5%).
Probably a locus distinct from cys-5, with the order leu-3 cys-5
(<1%)
cys-11
mt in crosses showing no negative interference (721, 723). (N.H. Horowitz,
cited in references 721 and 815)
Uses sulfite, thiosulfate, cysteine, or methionine (721). Affects adenosine 5'-triphosphate sulfurylase (639; F.-J. Leinweber, cited in references 721 and 723). Enzymatically distinct from cys-5 (3'-phosphoadenosine-5'- phosphosulfate reductase), which it complements in heterokaryons (721, 723; F.-J. Leinweber, cited in references 721 and 723). Called cys(NM86) (721). The cys(85518) mutant also lacks adenosine 5'-triphosphate sulfurylase (639), and thus cys(85518) is evidently an allele of cys-11 rather than of cys-5; this is in harmony with the existing recombination data (723).
cys-12: cysteine-12
IR. Right of ad-9 (12%); linked to al (0/76) (723).
Uses cysteine or methionine (723). No information on precursors used.
cys-13: cysteine-13
IR. Right of his-3 (2%) (640).
Resistant to chromate; no demonstrable requirement. Deficient in sulfur permease I (conidial type) (639, 640). Score on minimal agar with 25 mM chromate and 0.25 mM methionine after 3 days or longer, 34°C. (Strains carrying regulatory gene cys-3 are also chromate resistant.)
cys-14: cysteine-14
IV. Linked to cot-1 (21%) (640).
Deficient sulfate transport in the mycelial stage, but sensitive to chromate. Lacks sulfur permease II (mycelial type) (639, 640). Double mutant cys-14;cys-13 cannot transport inorganic sulfate, grows on methionine; both single mutants are prototrophic (640).
cys-15: cysteine-15
IVR. Between the T(S1229) breakpoints; hence, right of pdx-1
(0/55 asci).
Left of met-1 (3%) (55, 768, 808).
Unable to use sulfate. Uses sulfite, thiosulfate, cysteine, or methionine (721, 768). Only one allele is known, with requirement not separated from a deficiency of D-amino acid oxidase (768); this is thought to be due to a closely linked coincident lesion. (See oxD for other D-amino acid oxidase mutants having no cysteine requirement.) Formerly called cys(oxD1).
cys(oxD1)
Changed to cys-15.
cyt: cytochrome
Cytochrome-deficient Mendelian mutations have been subdivided into three
main classes as follows (87): cyt, deficiency of more than one cytochrome;
cya,
deficiency of cytochrome aa3; cyb, deficiency of cytochrome
b. Apart from
diagnostic spectra, the mutants are characterized by slow growth (4 to 7 days
for conidiation of mutants versus 3 days for the wild type) and by the inability
to reduce tetrazolium (87). These properties have been used in screening new
mutants. cni also affects cytochrome spectra (309). The relation of
tet
(tetrazolium-resistant nonreducer) to cytochrome mutations is not known.
Cytochrome defects that result from mutations of the mitochondrial genome
(e.g., [mi-1] and [mi-3]) are not considered here except as they
interact with
chromosomal genes such as su([mi-1]); see reference 394.
cyt-1: cytochrome-1
IL. Between leu-3 (5 to 8%) (583) and T(OY321) (D.D. Perkins,
N.B. Raju,
and E.G. Barry, in preparation). (694)
Deficient in cytochromes aa3 and b. Very slow growth. Female sterile (87, 694). Scoring aided by slower growth on complete medium relative to that on minimal, presumably owing to inhibition by yeast extract (694, 816). cyt-U-9 is linked and may be allelic with the original cyt-1 mutation C115 (87; H. Bertrand, personal communication).
cyt-2: cytochrome-2
VIL. Between cya-3 (10%) and lys-5 (6%) (87, 1012).
(694)
Completely deficient in cytochromes aa3 and c. Very slow growth. Female sterile. Complements cyb-2. (87, 694) Spectrum (87). Cytochrome oxidase subunit 1 polypeptide abnormal (90). aa3 deficiency suppressed by cyb-1 and by antimycin A (84).
cyt-3: cytochrome-3
The original mutation cyt-3-5 (87) is evidently an allele of cyt-4 (H.
Bertrand,
personal communication).
cyt-4: cytochrome-4
IR. Between the breakpoints of T(AR173); hence, right of the centromere,
sn,
and arg-3 (2%); left of nuc-1 and lys-4 (808; D.D.
Perkins, unpublished data).
(87)
Deficient in cytochromes aa3 and b. Slow growth. cyt-4-7
and strains carrying
the three alleles listed below are all defective in splicing of the large
mitochondrial rRNA (83; H. Bertrand, personal communication). Phenotype
partially suppressed by the electron transport inhibitor antimycin (83).
Complements cyt-18 and cyt-19, which are also required for the
splicing of
large mitochondrial rRNA (83). cyt-3-5, cyt-U-10, and cyt-U-14 of
reference 87
are all alleles of cyt-4 and have been designated cyt-4-5, cyt4-10,
and cyt-4-14,
respectively, by H. Bertrand (personal communication).
cyt-5: cytochrome-5
IVR. Left of trp-4 (9%) (87).
Deficient in cytochromes aa3 and b. Slow growth. Not defective in the splicing of mitochondrial rRNA, unlike cyt-19 mutants (cyt-19 is closely linked but complements cyt-5) (83, 87). Allele cyt-5-4 was previously called cyt-4.
cyt-6: cytochrome-6
VII. Near wc-1 (2%). Indicated to the left (87)
Deficient in cytochromes aa3 and b. Slow growth. cyt-U-18 may be an allele (0/193 cyt+ from a cross with cyt-6; positive complementation test). Not tested for allelism with slo-2. (87)
cyt-7: cytochrome-7
VIIL. Linked to nic-3 (18%). Indicated to left (87).
Deficient in cytochromes aa3 and b. Slow growth. Possible allele of su([mi-1])-5. (87)
cyt-8: cytochrome-8
IIIR. Linked near ad-4 (87).
Deficient in cytochromes aa3 and b. Possibile allele of su([mi-1])-1. (87)
cyt-9: cytochrome-9
V. Between lys-1 (5%) and at (5%) (87, PB).
Deficient in cytochromes aa3 and b. Slow growth. (87)
cyt-12: cytochrome-12
IIR. Between thr-3 (38%) and trp-3 (18%). (87)
Incompletely deficient for cytochromes aa3 and c. Slow growth. Female sterile. Complements cya-4. Spectrum. (87)
cyt-18: cytochrome-18
IR. Linked to al-2 (10%) and nic-1 (1 to 5%) (203, 635).
Heat sensitive; grows slowly. Deficient in cytochromes aa3 and b at 37°C (832). Mitochondrial protein synthesis and assembly of small mitochondrial subunits are also abnormal (205). A novel large RNA precursor (35S) is found at restrictive temperatures (205); the intervening sequence of large (25S) mitochondrial rRNA is apparently not excised. Two alleles (289-67, 299-9) differ in speed of turn-off of RNA processing when the temperature is shifted (635). Complements cyt-4 and cyt-19, which are also required for splicing of large mitochondrial rRNA (83).
cyt-19: cytochrome-19
IVR. Linked to cyt-5 (1/201) and trp-4 (9%) (83, 87).
Deficient in cytochromes aa3 and b. Slow growth. Required for splicing of mitochondrial large rRNA. Complements cyt-5. Complements cyt-4 and cyt-18, which are also required for splicing (93). Called cyt-U-19 (87).
cyt(289-56): cytochrome
IL. Linked to mt (0/151) (203).
Deficient in the small subunit of mitochondrial ribosomes, but contains normal ratios of 19S to 25S rRNA in whole mitochondria (205).
cyt(297-24): cytochrome
II. Linked to thr-3 (33%), probably to the left (203).
Deficient in the small subunit of mitochondrial ribosomes. 19S rRNA is rapidly degraded. The phenotype resembles that of the extranuclear [poky] mutant (205), but the deficiency is not suppressed by su([mi-1])-4 or su([mi-1])-5 (203).
Last modified 4/24/96 KMC