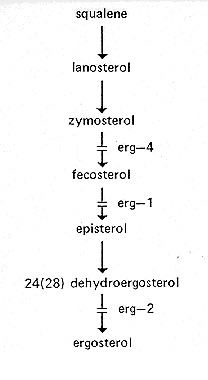
FIG. 13. Probable pathway of sterol biosynthesis, showing sites of gene action (419, 699, and references therein).
Easily Wettable (eas)
Ergosterol (erg)
Female Fertility (ff)
Fluffy (fl)
Fluorophenylalanine Resistant (fpr)
Frequency (frq)
Glycerol phosphate (glp)
Gulliver (gul)
d
Used as symbol for het-d, q.v.
da: dapple
IIL. Linked to thr-3 (3%) and arg-5 (812, 818). Right
of T(NMI49) (D.D. Perkins,
unpublished data).
Produces flecks of conidia on the agar surface (812).
del: delicate
VIR. Right of rib-1 (12%) and pan-2 (6%). Left of trp-2
(0 to 13%) (818). (789)
Growth less profuse than that of the wild type; flat, with fine aerial growth at the tops of slants. Prolific hyphal branching. Scorability good. (789)
dgr: deoxyglucose resistant
V. Linked to caf-1 (8%) (B.M. Eberhart, personal
communication).
Resistant to inhibition by 2-deoxy-D-glucose. Grows more slowly than the wild type on standard media, but growth is initially faster than that of the wild type on media with mono- or disaccharides plus deoxyglucose. The greatest differential growth in 0.05% deoxyglucose is obtained with 0.1% cellobiose, trehalose, lactose, fructose, or galactose. (298)
dir: dirty
IR. Right of pa (37%) (610).
Conidia few and misshapen; yellowish exudate (611). Photograph (610). (Stock lost. Possibly os-1?)
dn: dingy
IVR. Right of pyr-2 (4%). Linked to mat (1%) (692,
812).
Abnormal morphology, slower than normal growth, producing grey patches of microconidia in addition to macroconidia (692). When in combination with fl, microconidia are produced exclusively and in abundance, as in the double mutant pe fl. The double mutant fl;dn is fully fertile in homozygous crosses (811) and for this reason may be preferred to pe fl as a microconidating strain. Microconidia from fl;dn are less viable, however (454).
do: doily
VIIL. Left of nic-3 (1 to 3%) (304, 812). Linked to spco-4
(1/>400) (816). (D.R.
Stadler, cited in reference 812)
Restricted colonial growth (D.R. Stadler, cited in reference 812); growth rate is 4% of the wild-type rate (304). The cell wall galactosamine is 0.5% the wild-type level, the uridine 5'-diphosphate N-acetylgalactosamine content is 3%, and the specific activity of uridine 5'-diphosphate N-acetylglucosamine-4-epimerase in cell extracts is 20% that of the wild type. Partial back-mutations can differentially affect cell wall and alcohol-soluble galactosamines, indicating pleiotropy (304). Cell walls have a reduced amount of peptides, and the peptides have altered diethylaminoethyl cellulose elution profiles (1165).
dot: dot
IR. Linked to ad-9 (0/44), right of thi-1 (2%) (816).
(812)
Colonial growth (812). More restricted on glycerol complete medium than on minimal medium. A possible "maternal effect" is seen in dot+ progeny from heterozygous crosses.
dow: downy
IIIR. Right of ty-1 (21%) and un-17 (23%); linked to
erg-3 (10%) and acu-7 (0/72) (816).
Soft, matty growth, conidiating and covering slants (816). An excellent marker: good viability, fertility, and scorability.
dr: drift
VIIR. Right of for (3%), left of T(5936) and arg-10
(12%) (808, 819, PB).
Forms conidia in dense masses at the tops of slants; growth elsewhere is flat on the surface. Good scorability. (819) Called mo(P1163).
e
Used as symbol for het-e, q.v.
eas: easily wettable
IIR. Linked to rip-1 (1/151), trp-3 (0/71), and fl (1/52)
(PB). (967)
Conidia and aerial hyphae are readily wetted by water; in contrast, those of wild type are hydrophobic. Resembles csp mutants in that conidia do not readily become airborne, but differs from csp mutants in that conidia do not remain joined in the proconidial chains (967). Rodlets are lacking from the surface of conidia (75). Conidiating cultures can be scored by adding a drop of water to the culture, by tapping an inverted slant (967), or by transferring conidia to liquid (PB). Not scorable in combination with fl. Somewhat sensitive to high osmotic pressure (PB). A class of slow-growing progeny is produced from crosses heterozygous or homozygous for eas (811).
edr-1: edeine resistant-1
VI. Linked to ad-1 and pan-2 (0/125) (1064).
Resistant to edeine. Selected and tested on 200 µg of edeine per ml (only a fraction of edr conidia grow). Recessive. Called edr-1 (1064).
edr-2: edeine resistant-2
VIL. Left of ad-1 (19%) (1064).
Resistant to edeine. Selected and tested on 200 µg of edeine per ml (only a fraction of edr conidia grow). Recessive. (1064) In intact cells, edeine inhibits the syntheses of protein, deoxyribonucleic acid (DNA), and RNA in the wild type but not in the mutant; in vitro, edeine inhibits protein synthesis equally in both the mutant and wild type. Hence, the mutant is thought to have a block in edeine uptake. (1110) Called edr-2, edr-29.
en(am)-1: enhancer-1 of am
VR. Between am (8%) and inl (1%). Linked to gln
(1%). (122, 339, 345)
In en(am)-1 am double mutants, en(am)-1 blocks the adaptation of am on minimal medium without a source of amino nitrogen (345). The double mutants are inhibited by ammonium and grow adequately only when glutamate is the sole nitrogen source. The en(am)-1 single mutant grows well on minimal medium, but is unable to use, as the sole nitrogen source, proline (122), methionine, alanine, isoleucine, valine, urocanate, hypoxanthine, uridine, urea, or bovine serum albumin (184). It is relatively resistant to p-fluorophenylalanine and ethionine and completely resistant to 0.02 M glycine. These properties cosegregated with en(am)-1 in all isolates tested (339). Glutamate synthase (GOGAT) is normal (293). The single mutant is scored by using minimal medium with proline as the sole nitrogen source (122) or (better) by using 0.2 mM p-fluorophenylalanine (339). Name changed from i (inhibitor) (293).
en(am)-2: enhancer-2 of am
IIR. Linked near pe (983).
In en(am)-2;am double mutants, en(am)-2 counteracts leakiness of am on minimal medium. en(am)-2;am strains grow well on L-alanine (25 mM) or 0.5% casein hydrolysate (983) or on glutamate (5 mM) (293). The single mutant en(am)-2 without am grows normally on minimal medium. Mutant en(am)-2 lacks glutamate synthase (GOGAT) (see Fig. 19). The double mutant en(am)-2;am lacks NADP-glutamate dehydrogenase and GOGAT activities (293). Frequent revertants of the double mutant en(am)-2;am on suboptimal medium are attributed to back-mutation at am (983). Formerly called en-am.
En(pdx): Enhancer of pdx-1 pigment
IL. Linked to mt (5%), probably to the left (637).
En(pdx);pdx-1 double mutants, grown on Vogel medium (1103) or on Westergaard and Mitchell medium (1134) supplemented with ammonium sulfate at 5 g/liter but not at 1 g/liter, excrete yellow pigment into the medium. This property is not shown by either single mutant (637). The addition of a nonlimiting concentration of pyridoxine inhibits production of the pigment on both media. Production of the pigment is also inhibited in heterokaryons between complementing pdx alleles. En is dominant over En+ in heterokaryons between noncomplementing pdx alleles (847).
er: erupt
Allelic to rg-1, q.v. (382). The symbol er-1 has been
abandoned.
erg: ergosterol
Ergosterol mutants have been detected by resistance to nystatin and other
polyene,antibiotics. The known mutants are female sterile. Intercrosses for allelism tests
can be made, however, by using a heterokaryon as the female parent (419, 699).
Ergosterol biosynthesis is illustrated in Fig. 13.
erg-1: ergosterol-1
VR. Between pk (2%) and asn (9%) (419).
The cell membrane is deficient in ergosterol, conferring strong resistance to nystatin and other polyene antibiotics (418, 419). Defective in conversion of fecosterol to episterol (Fig. 13.). Lacks fecosterol delta8,delta7-isomerase (419). Infertile as female. Slow growth, reduced conidiation. The nysr mutants of reference 699 are blocked in the same reaction, but nysr has not been tested for allelism with erg-1.
erg-2: ergosterol-2
VR. Left of inl (6%) (419).
The cell membrane is deficient in ergosterol, thereby conferring slight resistance to nystatin and other polyene antibiotics (417, 418). Lacks 24(28)dehydroergosterol hydrogenase (terminal step of ergosterol synthesis) (419) (Fig. 13.). Poorly fertile as female. Good growth and conidiation.
erg-3: ergosterol-3
IIIR. Linked to dow (10 to 14%), probably to the left (PB).
The cell membrane is deficient in ergosterol; slightly (2x) increased resistance to nystatin and other polyene antibiotics (417, 418). Biosynthetic lesion not identified (419). Female sterile, but forms tiny protoperithecia. Slow growth, reduced conidiation, uneven production of aerial hyphae.
erg-4: ergosterol-4
IR. Linked to al-1 (10%) (PB).
The cell membrane is deficient in ergosterol, conferring slight resistance to nystatin. Lacks C24 (zymosterol)-methyl transferase. Accumulates zymosterol (Fig. 13.). Infertile as female (419). Slow growth, colonial at 34 C, spreading at 25°C (PB)

FIG. 13. Probable pathway of sterol biosynthesis, showing sites of gene
action (419,
699, and references therein).
eth-1: ethionine resistant
IL. Between arg-1 and arg-3 (1%) (816). (672)
Resistant to ethionine at 24°C (672). Labile S-adenosylmethionine synthetase (515, 547) (see Fig. 17). Resistance attributed to overproduction of methionine (541, 542). Heat sensitive; no growth at 37°C (672). Levels of several enzymes that are normally repressible by methionine are not repressed by methionine in eth-1R strains even at the growth-permissive temperature (124, 664). See also reference 965. Both heat sensitivity and ethionine resistance are reparable by high osmotic pressure (664). Called r-eth-1.
exo-1: exoamylase-1
I. Linked to mt (7%) and ad-3A (22%). Stated left of
mt, but data not given (325).
Hyperproduction of beta-amylase, alpha-amylase, glycoamylase, invertase (beta-fructofuranosidase), and (to a lesser extent) trehalase (404, 405, 1027). Enzymes secreted abundantly on depletion of exogenous carbon source (404). A polysaccharide is also released (325). Sevenfold increase in conidial enzyme levels. Altered amino sugar content of cell wall (404, 405). Initial allele called SF26, exoa-1 (325, 404). Probable second allele found in a strain of inl (89601) a (194, 325, 1027); allelism evidence in reference 325. With SF26, high amylase and high invertase levels cosegregated in 91 isolates (405). With allele from the 89601 strain, in a mixed-background cross, high amylase and high invertase each act as if due to a single major gene with many modifiers; high amylase and high invertase usually cosegregate and are not correlated with alkaline phosphatase levels (1027). The relation of exo-1 to gene VI-178, which reverses repression of invertase and trehalase production by mannose, is not known (663). For a linked gene defective in glycoamylase, see sor(T9) (called gla in reference 50 and called amy in reference 325). exo-1 has not been tested for allelism with sor(T9), but has been stated to be on the opposite side of mt. Because inl (89601) a has been used to obtain mutants by inositol-less death enrichment, exo-1 may be present but unrecognized in many laboratory stocks.
f: fast
See su([mi-1])-f
fas: fatty acid synthesis
See cel.
fdu-1: fluorodeoxyuridine resistant-1
Allelic with ud-1, q.v. (126).
fdu-2: fluorodeoxyuridine resistant-2
IV. Right of cys-4 (2%) (126, 463).
Resistant to 5-fluorodeoxyuridine, 5-fluorouracil, and 5-fluorouridine. Resistance is partially dominant in heterokaryons. Involved in regulation of pyr-3, udk, and ud-1. (127) Scored by spotting a conidial suspension on medium containing 4 x 10(-5) M filter-sterilized 5-fluorodeoxyuridine (463).
female fertility
Infertile as the female, but fully fertile as the male (fertilizing), parent. Besides
those listed, 32 more ff mutations have been obtained (530), but they are not
listed
separately because they are unmapped and lack locus numbers. These comprise 28
complementation groups and have been characterized with respect to position of
the block in perithecial development, dominance, effects on vegetative growth,
supplementability, and independence of mating type (530). These mutants are available
from the FGSC. Another symbol, fs (female sterile), has been used by other
workers for
mutant genes causing the same phenotype. Additional mutations, including some that
specify impaired female fertility as part of a pleiotropic syndrome and have other names,
are listed under fs. For additional characteristics, see fs.
ff-1: female fertility-1
IIR. Between aro-1 (5%) and un-20 (4%) (1052).
(1053)
Female sterile with no protoperithecia (1052, 1053). Enhanced glycerol utilization; aconidial on liquid glycerol medium. Good conidiation, but reduced aerial hyphae and heavy surface growth on sucrose minimal slants. Growth on glycerol reduces levels of both pyruvate dehydrogenase and dihydrolipoyl transacetylase (211). Effect of carbon source studied (1078). Conveniently scored by failure to produce protoperithecia on small slants of synthetic cross medium (7 days, 25 C). Allele T30 was originally recognized as specifying female sterility and was called ff-1 (1052, 1053). Allele JC744 was first characterized by glycerol utilization and called glp-3 (211).
ff-2: female fertility-2
Unmapped.
No protoperithecia. Normal vegetative morphology (459).
ff-3: female fertility-3
IR. Right of os-1 (3%) (193).
Defective in protoperithecial production. Abnormal morphology. Found in strain T22, which also contains ty-3 and ty-4. Not alielic to ty-3 or T. (193, 460) (Possible allele of so?)
ff-5: female fertility-5
IIIR. Between pro-1 (2%) and met-8 (1%) (1052).
Produces sterile brown protoperithecia without trichogynes and darkens medium (photograph). Vegetatively normal. Closely resembles ff-6. Not allelic to ty-1 or ty-2. (1052)
ff-6: female fertility-6
IIIR. Linked near ty-1 (459).
Produces many large black protoperithecia, but no perithecia are formed when used as the female. Black pigment excreted into medium (459).
fi: fissure
IV. Between cut (4 to 10%,) and pyr-1 (12 to 19%) (PB). Right
of
ace-4 (10 to 17%)
(578). (812)
Produces exudate in fissures formed under the agar and on the surface. Variable expression; may be difficult to score for some isolates (578, 812). Best scored on minimal synthetic cross medium, 34 C, pH 6 (78).
fl: fluffy
IIR. Between ace-1 (5 to 11%) and trp-3 (3%) (816, PB).
(613)
No macroconidia (609). Highly fertile (612). Used routinely as the female parent in tests for chromosome rearrangements and for mating type (e.g., reference 801). The fl single mutant produces few microconidia when dry; when wetted, sufficient microconidia are produced to have been used in early irradiation and mutation studies (614, 915); large numbers can be obtained under certain conditions; see reference 893. pe fl (46, 700) and fl;dn (806) double mutants produce abundant microconidia; the latter combination is highly fertile when homozygous. Photograph of microconidial formation (774); see also reference 893. Nuclear numbers in microconidia (46, 64, 478). Wall analysis (207). Immunoelectrophoretic pattern (784). Paradoxical high alcoholic glycolysis on nitrate medium (80). Deficiency of isocitrate lyase on acetate medium; see citations in reference 1088. When fl A and fl a strains are inoculated separately on crossing medium in plates, a double line of perithecia forms where they meet, similar to that accompanying barrage in Podospora (410, 414). fl ascospores from certain fl x fl+ crosses often germinate spontaneously (1127; N. B. Raju, personal communication). Allele C-1835 was called acon (717, 812).
fld. fluffyoid
IVR. Left of his-5 (2%) (991). (812)
Resembles fluffy mutants, producing no macroconidia (812).
flm-1: flame-1
Allelic with os-1, q.v.
flm-2: flame-2
Allelic with os-4, q.v.
fls: fluffyish
IR. Between nit-1 (5 to 19%) and al-1 (6 to
19%) (P. St. Lawrence, cited in references 47,
789, or 812).
Morphology of young cultures resembles that of fluffy; later conidiates. Suboptimal growth response to methionine (789). Called un(STL6).
fmf-1: female and male fertility
I. Between mt (2 to 15%) and cr-1 (2%).
Linked to arg-1 (531).
Perithecial development is blocked 15 h after fertilization, before meiosis, when fmf-1 is present either in the female or male parent. Perithecia attain only 40% normal diameter. Recessive in heterokaryons and can be crossed as one component of a heterokaryon, either as female or as male. Female fertility is also restored in mixed mating type (fmf-1 A + fmf+a) heterokaryons that are homokaryotic for tol (531). Called PBJ6 (527).
for: formate
VIIR. Right of wc-1 (5%) and frq (3%). Left of
dr (3%) (812, 819; J. F. Feldman, personal communication).
Requires formate or formaldehyde. Growth aided slightly if glycine, histidine, or choline is added to formate (446). Will also grow on a mixture of methionine and adenine and suboptimally on adenine alone (446). Lacks cytosolic (but not mitochondrial) serine hydroxymethyl- transferase (Fig. 17) (124, 210). Has increased formyl tetrahydrofolate synthetase, methylene tetrahydrofolate dehydrogenase, isocitrate lyase, and glyoxalate aminotransferase activities (209, 210). Use 0.3 mg of formate per ml. Can be autoclaved.
fpr: fluorophenylalanine resistant
Many mutants isolated by resistance to p-fluorophenylalanine are actually bradytrophs
(very leaky auxotrophs); most require one of
many amino acids. The resistance disappears
when the required growth factor is added. It is
suggested that the resistance is due to transinhibition of amino acid transport caused by
increased amino acid pools resulting from deprivation of the required amino acid. (552
and references therein) It is not clear whether any of the
mapped fpr mutations are of this type.
fpr-1: fluorophenylalanine resistant-1
VR. Linked to cyh-2 (<1%) (555, 1149).
Resistant to p-fluorophenylalanine and 4-methyltryptophan. Isolated as resistant to p-fluorophenylalanine in the presence of su(mtr)-1. Resistance is recessive in heterokaryons; used in mutagenicity test system (626). Suppressed by several lys and arg auxotrophic genes, which apparently give the double mutant greater sensitivity to p-fluorophenylalanine with no increase in uptake (555). Scored on solid medium containing 10 µg of p-fluorophenylalanine or 60 µg of 4-methyltryptophan per ml; see reference 550.
fpr-3: fluorophenylalanine resistant-3
IIIR. Linked to trp-1 (0.35%) and thi-2 (5%)
(550).
Resistant to p-fluorophenylalanine but not to 4-methyltryptophan. Not resistant in the presence of indole. Amino acid uptake is normal through transport systems I and II, as defined in reference 550. Isolated in a su(mtr) strain. Scored on solid medium containing 10 µg of p-fluorophenylalanine per ml (550), added before autoclaving.
fpr-4: fluorophenylalanine resistant-4
VR. Right of inl (11%) (550).
Resistant to p-fluorophenylalanine and 4- methyltryptophan. isolated in a su(mtr) strain. Not tested for amino acid uptake. Scored on solid medium containing 10 µg of p-fluorophenylalanine or 60 µg of 4-methyltryptophan per ml. (550)
fpr-5: fluorophenylalanine resistant-5
1. Left of al-2 (25%) (550).
Resistant to p-fluorophenylalanine, but not to 4-methyltryptophan. Isolated in the wild type. Not tested for amino acid uptake. Scored on solid medium containing 10 µg of p-fluorophenylalanine or 60 µg of 4-methyltryptophan per ml. (550)
fpr-6: fluorophenylalanine resistant-6
VIR. Between pan-2 and trp-2 (247).
Resistant to p-fluorophenylalanine. The only allele, UM300, was found in a variant unable to take up arginine to satisfy requirement of arg mutations. This blockage is manifest mainly when ammonium is in the medium. Uptake of many other metabolites (amino acids, uridine, sugars) is also affected. Primary defect unknown (247; R. H. Davis, personal communication). Called UM300 or fpr(UM300). Not tested for allelism with mts or mod-5, which map in the same area and cause increased rather than decreased uptake.
fr: frost
IL. Between ro-10 (18%) and un-5 (6%) (798,
PB). (789)
Delicate branching on agar surface and delicate aerial growth with no conidia (789). Multiple hyphal branching (382). Deficient in glucose- 6-phosphate dehydrogenase (as are col-2 and bal mutants) (949, 952). Partially deficient in linolenic acid (115); morphology partially corrected by exogenous linolenic acid (892, 943). Low adenylate cyclase activity and low adenosine 3',5'-phosphate (943, 950). Used to determine what functions are controlled by adenosine 3',5'-phosphate (779). Unlike cr-1, fr is not corrected morphologically by exogenous cyclic nucleotides (892, 951). Scott (943) reported that morphology is corrected by theophylline; Rosenberg and Pall (892) reported no correction by phosphodiesterase inhibitors. Cell wall analysis; photograph (112, 278, 946). Reduced amount of cell wall peptides (1165). Recessive in duplications (808). Female sterile. Both known alleles (B110 and R2499) revert to fr+.
frq: frequency
VIIR. Between un-10 (9%) and for (3%) (J.F.
Feldman, personal communication). Right of
met-9 (9 to 13%). Linked to oli (< 2%); possibly
allelic (282). (330)
A series of clustered genes or of multiple alleles resulting in altered periods in the circadian rhythm cycle of conidiation. Identified mutations and their periods (at 25 C without csp) are: frq-1, 16.5 h; frq-2,19.3 h; frq-3, 24.0 h; and frq-4, 19.3 h (331); frq-6, 19.2 h; frq-7, 29.0 h; and frq-8, 29.0 h (wild type, 21.5 h) (329, 377). Strains carrying allele UV-111-9 show erratic periodicity. Temperature compensation and interactions with other loci have been described; dominance is incomplete; growth rates are normal (326, 375-377). Scoring is accomplished by zonation in growth tubes or plates, using strains that carry bd and preferably csp-1 or csp-2. Presence of csp shortened period length about 1 h in the one strain tested (279). Period-altering mutations elsewhere in the genome are given different names (e.g., prd and chr). The symbol frq is reserved for this locus or region (329). For review of circadian mutants, see references 326 and 328.
frq-5: frequency-5
Changed to prd-1, q.v.
fs: female sterile
Infertile as the female but fully fertile as the
male (fertilizing) parent. No or few functional
perithecia are produced. Another symbol, ff
(female fertility, q.v.), has been used by other
workers for mutants having the same phenotype. Female fertility is also impaired or
absent
in some mutants that were named for other
traits, e.g., cyt-1, cyt-2, erg, fr, glp-3, gul-3, gul-
4, leu-1, R, ro, sk, so, ssu, ty-1, ty-2, var-1.
Numerous additional female-sterile mutants
have been isolated (91, 253, 491, 530, and references cited therein), but the genes have
not been mapped and/or tested for allelism with the mutations listed here. Many of
these also affect vegetative morphology or growth rate. Tests on ff mutants,
q.v., show that different mutants are
blocked at different points in perithecial development (530). Female sterility has no
genetic or
functional relationship to mating type (530).
Crosses homozygous for any fs gene listed can
be made, and progeny can be obtained, by using
a heterokaryon of marked fs and fs+ strains as
the female parent (732; O. M. Mylyk, personal
communication); the same is true for most ff
genes (530).
The term "sterile" has been used in different
ways: for situations in which no protoperithecia
are formed, or in which perithecium development is blocked before ascospore
formation, or
even in which ascospores are produced that are
inviable. The term "barren" has been proposed
specifically for crosses in which perithecia develop but few or no ascospores are
produced
(860). See also ff and pp.
fs-1: female sterile-1
I or II. Linked to T(I;II)4637. Unlinked to mt
(732).
Perithecia are absent or infrequent when used as the female (protoperithecial) parent. Fertile as the male. Some strains produce occasional perithecia and ascospores. Vegetative growth is somewhat stringy, slightly slower, and paler than that of the wild type. Recessive in heterokaryons. Complements fs-2, -3, 4, -5, -6, and -n. (732; O.M. Mylyk, personal communication). Shown nonallelic with fs-2, -3, -5, and -n in crosses (732).
fs-2: female sterile-2
Tentatively II; probable loose linkage to fs-1 (732) and
cot-5 (14/48) (PB).
No perithecia when used as the female. Fertile as the male. Abnormal morphology, somewhat colonial. Grows at 25 C, but not at 34°C. Recessive in heterokaryons. Complements fs-1, -3, 4, -5, -6 , and -n. (O.M. Mylyk, personal communication)
fs-3: female sterile-3
IL. Left of mt (16%) (732).
No perithecia when used as the female. Fertile as the male. Vegetative growth is slightly slower and paler than that of the wild type. Recessive in heterokaryons. Complements fs-1, -2, 4, -5, -6, and -n (732; O.M. Mylyk, personal communication).
fs-4: female sterile-4
I? Linked mating type (22%) (732). May be
inseparable from a chromosome rearrangement
(O.M. Mylyk, personal communication).
No perithecia when used as the female. Fertile as the male. Complements fs-1, -2, -3, -5, -6, and -n. Vegetative growth is slightly slower and paler than that of the wild type. Recessive in heterokaryons. (732; O.M. Mylyk, personal communication)
fs-5: female sterile-5
I or II. Probable loose linkage with fs-1 (732).
Perithecia absent or rare when used as the female; some strains produce occasional perithecia and ascospores. Fertile as the male. Slow growth, mostly aerial near the surface of the agar. Cultures turn brown with age (732). Complements fs-1, -2, -3, 4, -6, and -n. (O.M. Mylyk, personal communication)
fs-6: female sterile-6
I or II. Linked to T(I;II)4637. Unlinked to mt
(732).
No perithecia when used as the female. Fertile as the male. Vegetative growth is slightly slower and paler than that of the wild type. Recessive in heterokaryons. Complements fs-1, -2, -3, 4, -5, and -n (732; O.M. Mylyk, personal communication).
fs-n: female sterile-n
1. Linked to mt (35 to 45%) and to T(I;II)4637
(732).
No perithecia when used as the female. Fertile as the male. Vegetative growth is slightly slower and paler than that of the wild type. Recessive in heterokaryons. Three complex ascus segregations in 51 asci suggest two closely linked genes. If so, they must not complement each other, although they complement fs-1, -2, -3, 4, -5, and -6. (732; O.M. Mylyk, personal communication)
Fsp-1: Four-spore-1
IIR. Right of pe (4%) (D.D. Perkins, unpublished data).
Some of the asci contain four large ascospores rather than the normal eight. In these asci, ascospores are formed at the four-nucleate stage after meiosis II. Dominant, with variable penetrance depending on genetic background. Ascospores from four-spored asci produce homokaryotic cultures. Rarely, three-spored or two- spored asci are formed, and these include some heterokaryotic ascospores. One postmeiotic mitosis is omitted in the four- and three-spored asci, and two divisions are omitted in the two-spored asci. Vegetative morphology is normal. (856) Used in the study of Sk (Spore killer) (857). The cytological basis of Fsp-1 is distinct from that in N. tetrasperma. For a description of a dominant eight-spored mutant of N. tetrasperma see reference 129.
Fsp-2: Four-spore-2
IR. Right of nic-2 (6%) (N.B. Raju, personal communication).
(253)
In crosses heterozygous for Fsp-2, nearly all asci are four-spored at 16°C and eight-spored at 25°C (253). Crosses homozygous for Fsp-2 make four-spored asci at both temperatures. The cytological basis is similar to that for Fsp-1 (N.B. Raju, personal communication).
fz: fuzzy
Unmapped.
Abnormal morphology; one component of the combination of mutant genes that results in the cell-wall-less "slime" phenotype (321).
G: gulliver
See gul-1.
gap: gap
IL. Between mt (6%) and the centromere (4%) (610).
Conidia in a few scattered clusters on long nonconidial hyphae. Photograph (610). (Stock lost.)
gla: glucoamylase
See sor(T9).
glm: glutamine
Changed to gln.
gln-1: glutamine-1
VR. Linked to inl (2%, probably to the right) (869).
Requires glutamine (869). Probably the glutamine synthetase structural gene (229, 912) (see Fig. 19). Mutants have altered enzyme (912). Sensitive to chlorate on both ammonium and glutamate; resistant to chlorate on glutamine (292). NADPH-nitrate reductase, NAD(P)H-nitrite reductase, and uricase are freed from repression by ammonium or glutamate but not glutamine in the gln-1a mutant (291, 294, 836, 1118). Allele gln-1b is more derepressed than allele R1015 (called gln-1a) (292, 837). For interaction with am, see reference 503. Formerly called glm (869).
glp: glycerol phosphate
Symbol replaces gly for mutants with altered
ability to use glycerol as a carbon source.
Scored on slants of minimal synthetic cross
medium (1134) with 2% glycerol versus 2%
sucrose as the carbon source (1102). Poor
growth of the wild type on glycerol medium is
markedly improved by the addition of 0.5% L-asparagine and 100 µg of ascorbic
acid
per ml
(189); this might facilitate testing. For diagram of
pathways of glycerol utilization in various organisms, see reference 1078 or 1102.
glp-1: glycerol phosphate-1
IR. Linked to ad-9 (2%) and nit-1 (11%);
probably between them (466, 763).
Unable to use glycerol as the sole carbon source (763). Can use dihydroxyacetone or glyceraldehyde (261). Probably regulatory. Deficient in inducible glycerol kinase under normal conditions (466, 764); wild-type levels of normal enzyme are induced by cold or by deoxyribose in strains carrying some, but not all, alleles (261, 466); glycerol transport is normal (261). Fine-structure map (262). Called gly and gly-u.
glp-2: glycerol phosphate-2
IIR. Right of T(ALS176); hence, of arg-5 (8%).
Left of T(NM177); hence, of pe (7%). Linked to
aro-3 (3%) and ff-1 (glp-3) (15%) (263).
Unable to use glycerol, dihydroxyacetone, or glyceraidehyde as the sole carbon source (261, 263). Lacks both mitochondrial and cytosolic flavin-linked glycerol-3-phosphate dehydrogenase (263). Three independent isolates all have altered ropy-like vegetative morphology, but are female fertile, unlike most ro mutants (212, 263). The report of complementation groups at this locus (214) is in error (J.B. Courtright, personal communication). Fine-structure map (262). Called gly-2.
glp-3: glycerol phosphate-3
Allelic with ff-1, q.v.
glp-4: glycerol phosphate-4
VI. Right of ad-1 (0 to 2%) and ylo-1 (1 to 6%). Left of
rib-1 (3 to 4%) and pan-2 (4
to 6%) (1102).
Unable to use glycerol as the sole carbon source (1102). Uses dihydroxyacetone or glyceraldehyde (261). Lacks both inducible and constitutive glycerol kinase (1102), but there is some doubt that these are two different enzymes (J.B. Courtright, personal communication, based on reference 262). Revertant with altered kinetic properties (262). Allele G660 originated in N. tetrasperma and was introgressed into N. crassa (77, 1102). Fine-structure map (262).
glp-5: glycerol phosphate-5
1. Left of cr-1 (15%) (1102).
Unable to use glycerol as the carbon source. Lacks glyceraldehyde kinase (1102), but the significance of this is uncertain because of findings reported in reference 1078. Allele M1051 originated in N. tetrasperma and was introgressed into N. crassa (77, 1102).
glp-6: glycerol phosphates
V. Left of inl (30%) (840).
Deficient in NAD-linked glycerol-3-phosphate dehydrogenase. Called 42-94 (840; H.B. Howe, Jr., personal communication).
glt: glycyl-leucyl-tyrosine resistant
Unmapped.
Unable to transport oligopeptides necessary to support growth of specified amino acid auxotrophs (1155). Has only 10% of the wild-type uptake rate (1154). (Oligopeptide uptake system transports tri-, tetra-, and pentapeptides, but not di- or higher than pentapeptides.) Obtained, using tys, by selecting mutants resistant to glycyl-L-leucyl-L-tyrosine but still sensitive to tyrosine (1155). See reference 1151 for a review of peptide uptake.
gluc-1: Beta-glucosidase-1
IIIR. Linked to dow (10%) (B.M. Eberhart,
personal communication).
Activity of the thermostable aryl-beta-glucosidase reduced to 10% of the wild type (300) by one allele, and to <1% in a second-step mutant then called gluc-2, which showed 0/200 recombination with the original mutation and is probably allelic (299). Low activity is dominant in heterokaryons (630). Selected by the p-nitrophenyl glucoside staining reaction (297). Scored by breakdown of the beta-glucoside esculin (0.01%) as measured by fluorescence at pH 5.5 (300), or by precipitation of ferric ammonium citrate (0.1%) by esculetin (2 days, 25 C) (B.M. Eberhart, personal communication).
gluc-2 See gluc-1.
gly or gly-u: glycerol utilization
Changed to glp.
gpi-1: glucosephosphate isomerase
IVR. Linked to ad-6 (10%) (711).
Lacks glucosephosphate isomerase (phosphohexoisomerase). Grows slowly and colonially on glucose or sucrose. Unable to use fructose, but growth on glucose is stimulated by added fructose. Growth is enhanced in double mutants with either sor(T9) or pp. Allele T66M37 was originally called gpi-2 (711).
gpi-2
See gpi-1.
gran: granular
VR. Linked to pl (0/75); between pab-2 (1 to
8%) and his-6 (8 to 27%) (816, PB). (812)
Delicate granular conidiation, with conidia adherent rather than powdery (812). Sparsely branched hyphae (382). Morphologically distinct from pl mutants. Reduced amount of cell wall peptides (1165).
grey: grey
IVR. Linked to cot-1 (4%) (499).
Produces grey conidia (microconidia?) in the presence of cr-1 (499). Attempts to obtain grey progeny have been unsuccessful (PB).
gs(3): gamma sensitive
Not mapped.
Sensitive to ionizing radiation, but not to UV. Normal UV-induced mutation. Evidence not given for nonallelism with gs(6) and gs(20) (662).
gs(6): gamma sensitive
Not mapped. (Perhaps VI.)
Sensitive to ionizing radiation and UV. De- creased UV-induced mutation. Evidence not given for nonallelism with gs(3) and gs(20) (662).
gs(20): gamma sensitive
Not mapped.
Sensitive to ionizing radiation but not to UV. Decreased UV-induced mutation (662).
gsp: giant spore
IL. Left of mating type (10%) (1008). (589)
Some asci contain a single giant ascospore; others have two very large ascospores or four double-size ascospores; some contain eight ascospores. Proportions of these types vary on different crossing media. Vegetative growth is weak with normal morphology. The giant ascospores have multiple germination pores. The mutant ascus phenotype is recessive (589, 1008).
gua-1: guanine-1
1. Linked to arg-3 (8%) (1171); probably between
his-2
(3%) and cr-1 (3%) (PB).
Requires guanine. Inhibited (competitively) by adenine and by complex complete medium. Adapts phenotypically after several days and grows up on minimal or complete medium, but retains the requirement on subculture. Adenine prevents or decreases adaptation. Guanosine is preferred to guanine as a supplement because of greater solubility. Best scored at 2 and 3 days on slants of minimal medium plus 1 mg of adenine versus minimal medium plus 0.2 mg of guanosine per ml. (1171) Deficient in inosine 5'-mono-phosphate dehydrogenase (10% of the wild-type level with allele OY301) (405a) (Fig. 8).
gua-2: guanine-2
IVR. Linked to cot-1 (5%) (405a).
Requires guanine. No inosine monophosphate dehydrogenase activity. Does not adapt to grow on minimal medium (405a) (Fig. 8).
gul: gulliver
Name given (875) to suppressors of cot-1,
which produce large colonies at restrictive temperatures at which unsuppressed
cot-1 mutants
make tiny colonies. Scorable in the presence of
cot-1 at 34 C, 2 days after transfer of small
inocula to solid medium.
gu1-1: gulliver-1
VR. Between am-1 (<0.01%; <1%) and ace-5
(<1%) (577, 998). (875)
Modifier of the colony size of the mutant cot-1 at restrictive temperatures (875, 1068). cot-1; gul-1 strains take 60 h to reach the stationary phase (32 to 34 C) compared with 12 h for cot-1 gul-1+ strains (1068). Female fertile with viable ascospores. Able to make heterokaryons. Of 36 independent gul mutations, 25 were gul-1 alleles (1068). Recombination within gul-1 is unaffected by rec-3, which acts on the nearby am-1 locus (998). Formerly called G (875).
gul-2: gulliver-2
Not mapped.
Phenotype similar to that of gul-1 strains (1068).
gul-3: gulliver-3
IVR. Linked to cot-1 (10%) and pyr-2 (7%)
(1068).
Modifier of the colony size of the mutant cot-1 at restrictive temperatures. Female sterile. gul- ascospores are black but inviable. Occasional gul- progeny arise from gul+/gul- pseudowild disomic ascospores. Unable to make heterokaryons (1068).
gul-4: gulliver-4
VII. Linked to nic-3 (17%) (1068).
Resembles gul-3 (1068).
gul-5: gulliver-5
VI. Linked to trp-2 (10%) (1068).
Modifier of the colony size of the mutant cot-1 at restrictive temperatures. Female fertile. gul- ascospores are black but inviable. (1068)
gul-6: gulliver-6
Not mapped. Unlinked to cot-1 (IVR), inl
(VR), nic-3 (VIIL), gul-5 (VI), or gul-2
(1068).
Said to resemble gul-3 (1068), but ascospore ripening and recovery from ascospores have been found to be good (PB).
Last modified 4/24/96 KMC