Neurospora mutants beginning with H through L
You may jump directly to the following sections:
Histidine (his)
Isoleucine (ile)
Inositol (inl)
Leucine (leu)
Lysine (lys)
has: hydroxamic acid sensitive
Not mapped. Unlinked to azs (311).
Lacks salicyl hydroxamic acid-sensitive respiratory pathway; cannot produce the
hydroxamate-sensitive respiratory pathway when grown in the presence of
chloramphenicol. Grows slowly in the presence of antimycin A (311).
Double mutant has;azs is unable to grow in the presence of antimycin A; the
wild type
and the has+;azs single mutant are able to grow well on antimycin
A. The double
mutant has;azs (called ANT-1: antimycin sensitive) was used to obtain
oligomycin-resistant (312) and succinate dehydrogenase-deficient (307) mutants. From
strain ANT-1, called alx-1 (308).
het: heterokaryon incompatibility
If two strains carry different alleles at one or more het loci, they are unable to
form
stable heterokaryons (378, 379). Protoplasmic killing occurs after fusion of unlike
hyphae (384) or after microinjection of cytoplasm or extracts into unlike strains (1145).
Photographs (384). When duplications (partial diploids) are heterozygous for
het alleles,
growth is inhibited and highly abnormal (761, 803). The incompatibility due to
het genes
is strictly vegetative; it does not reduce fertility. Ten het loci have been
identified, and
various others are inferred to exist (729). het-c, -d, -e, and -i were
first defined by
heterokaryon tests. The remainder (het-5 through -10) were
detected by using
duplications. The mating type alleles A and a also act as
het genes in N. crassa (66, 384,
761, 830), although some slow heterokaryotic growth may occur (422). Microinjection
experiments implicate proteins in the killing reaction (1138, 1145). Review and literature
citations: 232, 803. Lindegren and Rockefeller wild types are het-C, het-D,
and het-E
(1144; J. F. Wilson, personal communication). St. Lawrence 74A and Oak Ridge wild
types are het-C, het-d, and het-e (1144). St. Lawrence 74A and
Oak Ridge OR8-la are
het-i (831). Differences at het loci are very common in natural
populations (730).
het-c: heterokaryon incompatibility-c
IIL. Left of pyr-4 (1%). Not included in duplications from
T(AR18) or T(P2869);
hence, right of cys-3 and het-6 (729, 808, PB). (378)
Stable heterokaryons are not formed by strains that are het-C +
het-c (378, 379);
strains carrying het-C/het-c duplications show inhibited "brown flat"
morphology,
spreading to cover the slant but not conidiating (794, 803). Putative multiple alleles,
suggested by abnormal duplication phenotypes when chromosomes from various natural
sources were tested, may be due instead to additional het loci in the segment
tested (729,
795). Photograph: see Fig. 3 of reference 729.
het-d: heterokaryon incompatibility-d
IIR. Right of fl (25%) (378). Included in duplications from
T(ALS176) (805).
Stable heterokaryons are not formed by het-D + het-d strains
(378, 379); strains
carrying hetD/het-d duplications show inhibited spreading growth on slants,
with fine
subsurface hyphae and no conidia. These are distinguishable from strains carrying
het-C/het-c duplications, which have a coarser texture (805).
het-e: heterokaryon incompatibility-e
VIIL. Left of nic-3 (28%) (1144). Included in duplications from
T(T54M501)
(803).
Killing reaction after fusion of het-E and het-e is more rapid
and severe, and growth
inhibition of strains carrying het-E/het-e duplications is more severe than for
strains
carrying incompatible combinations of het-c or of mating type alleles (803,
1144).
Photograph of heterozygous duplication colony (729).
het-i: heterokaryon incompatibility-i
I or II. Linked to T(IR;IIR)4637 al-1 (831).
Recognized by cessation of growth of forced heterokaryons under certain conditions.
het-I nuclei eliminate het-i if initial frequency exceeds 30%
het-I. When more than 80%
are het-i, growth of a forced heterokaryon can continue without a change of
ratio.
Called I and i. (831)
het-5: heterokaryon incompatibility-5
IR. Between T(NM103) and the tip; hence, right of thi-1 (729).
Vegetative incompatibility recognized by inhibited duplications and subsequently
confirmed by heterokaryon tests (729, 730).
het-6: heterokaryon incompatibility-6
IIL. Included in duplications from T(AR18), hence, right of
cys-3 and left of het-c
and pyr-4 (729).
Vegetative incompatibility recognized by inhibited duplications from translocations
AR18, P2869, and NM149. No heterokaryon tests (729).
het-7: heterokaryon incompatibility-7
IIR. Between T(D305) and the tip; hence, right of ro-2
(729).
Vegetative incompatibility recognized by inhibited duplications from
T(D305). No
heterokaryon tests. (729)
het-8: heterokaryon incompatibility-8
VIL. Between chol-2 (19%) and ad-8 (12%) (729,
730).
Vegetative incompatibility recognized by inhibited duplications from
T(T39M777)
and subsequently confirmed by heterokaryon tests (729, 730). Photograph of
heterozygous duplication colony (729).
het-9: heterokaryon incompatibility-9
VIR. Between T(AR209) and the right tip (729).
Vegetative incompatibility recognized by inhibited duplications from
T(AR209).
Photograph of heterozygous duplication colony. (729)
het-10: heterokaryon incompatibility-10
VIIR. Between T(5936) and the tip; hence, right of dr
(729).
Recognized on the basis of inhibited duplications from T(5936).
Photograph of
heterozygous duplication colony (729).
hgu-4: histidylglycine uptake-4
VR. Between cyh-2 (7%) and ure-2 (10%) (1149).
(1153)
Cannot use L-histidylglycine to support growth of his-6 mutants (1153).
Reduced
approximately 50% in transport of most amino acids tested. Resistant to many amino
acid analogs (1149).
his: histidine
Most histidine auxotrophs are inhibited by complex media or by certain combinations
of amino acids with which histidine does not compete effectively for permeases of the
basic, neutral, and general amino acid transport systems. A histidine mutant can grow
on minimal medium plus histidine in the presence of either a basic amino acid or a
competing neutral amino acid, but not in the presence of both (434, 628, 646).
Histidine mutants were not obtained in early mutant hunts in which complex media were
used, but were recovered on histidine-supplemented minimal medium (434, 595). For
general studies, see references 162, 434, and 1123. For details of histidine biosynthesis,
see Fig. 14. Enzymes of histidine biosynthesis are derepressed coordinately with those of
tryptophan, arginine, and lysine (137, 1131); reviewed in reference 642. See
cpc-1.
Called hist.
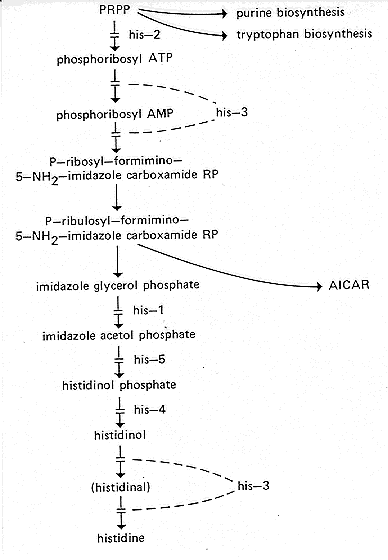
FIG. 14. Biosynthetic pathway of histidine, showing the sites of gene
action (16, 24,
25, 162, 673, 1123). ATP, Adenosine 5'-triphosphate; AMP,
adenosine 5'monophosphate;
PRPP, 5-phosphoribosyl pyrophosphate. AICAR
(5'-phosphoribosyl-5-aminoimidazole-
4-carboxamide ribosylphosphate) is an intermediate in purine synthesis. See Fig. 8. For
relations between histidine and purine synthesis, see reference 786.
his-1: histidine-1
VR. Right of ure-1 (1%). Left of pho-2 (3%), al-3,
and inl (1 to 10%) (397, 570, 578,
1036). (434)
Requires histidine (434). Accumulates imidazole glycerol phosphate. Lacks
imidazole glycerol phosphate dehydrase (24, 25) (Fig. 14).
Intralocus complementation
(162). Recombination between his-1 alleles is controlled by rec-1
(172, 520, 1070).
Initial his-1 allele called C84.
his-2: histidine-2
IR. Right of T(AR190) and un-2 (<1%). Left of the
T(AR173) right breakpoint and
of nuc-1 (<1%) (172, 670, 808). (434)
Requires histidine (434). Affects adenosine 5'-triphosphate
phosphoribosylpyrophosphate pyrophosphorylase (16) (Fig. 14).
Intralocus
complementation (162). Recombination between his-2 alleles is controlled by
rec-3
(173); it is not affected by rec-1 (172). Initial his-2 allele called
C94.
his-3: histidine-3
IR. Right of met-10 (R.L. Metzenberg, personal communication). Left of
cog (1 to 3%)
(172,174), ure-4 (1%) (78), and ad-3A (1%) (271).
(434)
Requires histidine (434). Complex gene coding for histidinol dehydrogenase,
phosphoribosyladenosine 5'-triphosphate-pyrophosphohydrolase, and
phosphoribosyl-adenosine 5'-monophosphate-cyclohydrolase (16, 673) (Fig. 14). All three
activities appear to be catalyzed by a single protein (673). Strains carrying different
individual alleles may lack only the early reaction(s) or only histidinol dehydrogenase, or
both. Those that lack only histidinol dehydrogenase accumulate histidinol (16, 162,
1123). Mutants produce cross-reacting material (220). Used to study intralocus
complementation and recombination (15, 16, 27, 162, 164, 171, 172, 1121, 1122, 1124).
Intralocus recombination is regulated by cog and by rec-2 (27, 171);
it is not affected by
rec-1 (172). Translocation T(IR;VII)TM429, with one breakpoint in
his-3, has been used
to show that cog is cis-acting (171). Initial alleles: C140 and T1710 (=
C1710).
his-4: histidine-4
IVR. Between cot-1 (1 to 4%) and met-5 (4%) (812).
(434)
Requires histidine (434). Accumulates L-histidinol phosphate. Lacks histidinol
phosphate phosphatase (24, 25) (Fig. 14). Allele P143h is heat
sensitive (his+ at 25 C);
C141 is not (815). Both are leaky (726).
his-.5: histidine-5
IVR. Between pyr-3 (1%) and trp-4 (3 to 7%) (991).
(162)
Requires histidine. Accumulates imidazole acetol phosphate and some imidazole
glycerol phosphate. Evidently lacks imidazole acetol phosphate transaminase (16, 162,
1123) (Fig. 14). Intralocus complementation (162) and
recombination (172).
his-6: histidine-6
VR. Right of un-9 (6%) and pyr-6 (6 to 18%). No recombination
with terminal
translocation T(NMI49) (793, 808, 816, 818, PB). Report of IV
linkage (646) not
confirmed.
Requires histidine. Blocked before imidazole glycerol phosphate (162, 1123) (Fig. 14).
No intralocus complementation (95 alleles) (162). Intralocus recombination (172).
his-7: histidine-7
IIIR. Between leu-1 (8 to 20%) and thi-2 (1 to 2%) (2199 1052,
PB). (162,
1123)
Requires histidine. Blocked before imidazole glycerol phosphate (162,
1123) (Fig. 14).
Intralocus recombination (172).
hist: histidine
Changed to his.
Histidine sensitive
Many mutagen-sensitive mutants are histidine sensitive (see mus, uvs, and
mei).
Several other histidine sensitives which are unmapped are not sensitive to the few
mutagens tested (see, e.g., reference 255); these are not listed pending evidence for
nonallelism with known loci.
hlp-1: histidinol permeability-1
VIIR. Between sfo (1 to 9%) and nt (28 to 37%). Left of
hlp-2 (8 to 25%)
(458).
Enables strains carrying a his-3 allele to use L-histidinol. This is proposed
to be due
to increased uptake through basic L-amino acid transport (system III as defined in
reference 778). The hlp-1 mutation confers increased sensitivity of
lys and arg mutants
to inhibition by arginine and lysine, respectively. (458)
hlp-2: histidinol permeability-2
VIIR. Between sfo (3 to 7%) and nt (29%). Right of
hlp-2 (8 to 25%) (458).
Enables a his-3 mutant to grow on L-histidinol. Growth of the double mutant
his-3;hlp-2
on histidine is inhibited by methionine, isoleucine, valine, and asparagine (458).
hom: homoserine
IR. Right of arg-6 (1%), Tp(T54M94), and al-2 (2 to
7%). Between the breakpoints of
T(STL76) and T(4637); hence, left of al-1 (cnr (1%), probably to the
left (789, 812). (1061, 1063)
Uses homoserine, or methionine plus threonine (1063). Affects aspartate
beta-semialdehyde dehydrogenase (518) (see Fig. 17).
Inhibited on complex complete
medium and by methionine and other amino acids (1063); supplemented minimal
medium should therefore be used. Symbol changed from hs.
hs: homoserine
Changed to hom.
i: (inhibitor)
Changed to en(am)-1.
i: (heterokaryon incompatibility-i)
See het-i.
i: (intensifier)
Used for unmapped intensifier of carotenoid pigment (982).
Iasc: Indurated ascus
VR. (A.M. Srb, personal communication)
Ascus wall hardens and darkens, so that the entire ascus resembles a giant ascospore.
Pores are formed and striations appear. Asci are germinable. Mutant ascus phenotype
is dominant with variable expression. Some asci are normal. Vegetative growth is weak
(1008). Resembles the indurated ascus phenotype described in N. tetrasperma
(285).
ile-1: isoleucine-1
VII. Between ars (1%) and wc-1 (3%). Probably right of
met-7 (<1 to 2%) (666,
PB). (812)
Uses isoleucine, alpha-amino-n-butyric acid, threonine (1061), or canavanine (63).
Affects threonine dehydratase (549, 552) (synonym: threonine deaminase [549]) (see Fig. 15). Leaky on minimal medium: treacherous to score with
large inocula. Tests should
be read early (24 h, 34 C). Moderate inhibition by methionine (1061). Selectable
as tiny germlings from ascospores germinated on minimal medium (666). Name changed
from thr-1 (549).
ilv: isoleucine plus valine
Three loci specify enzymes that catalyze corresponding steps in the parallel
biosynthetic pathways of isoleucine and valine (Fig. 15). These
enzymes are located in
the mitochondria (79, 597 and references therein) and may indirectly affect electron
transport (79). The enzymes may be in an aggregate; for a review, see reference 237.
The requirements are for both amino acids. A ratio of 20-30% isoleucine to 80-70%
valine is optimal (99). At least some ilv mutants are inhibited by norleucine,
norvaline,
phenylalanine (99), or tryptophan (J.F. Leslie, personal communication).
Enzyme production in response to end-product-derived signals depends on the
leu-3+ product and alpha-isopropylmalate. In leu-3+ strains,
threonine deaminase
production is repressed as a function of available isoleucine, acetohydroxyacid synthetase
is repressed as a function of valine, and isomeroreductase and dihydroxyacid dehydratase
are repressed as a function of isoleucine and leucine. In the absence of effective
leu-3
product, alpha-IPM, or both, enzyme production is repressed even under severe end
product limitation. (771) Formerly called iv.

FIG. 15. Biosynthetic pathways of isoleucine, valine, and leucine,
showing sites of
gene action (22, 136. 426, 549, 727. 854, 1112). Isoleucine and valine are synthesized
along parallel pathways catalyzed by common enzymes. The leucine precursors alpha-
and beta-isopropylmalate were formerly called beta-OH-beta-carboxyisocaproate
and alpha-OH-beta-carboxyisocaproate, respectively.
ilv-1: isoleucine plus valine-1
VR. Between per-1 (4%) and lys-2 (4 to 7%) (3, 14, 489). Left of
ilv-2 (<1 to 9%
prototrophs; some crosses give anomalous high frequencies [557]). (482)
Requires both isoleucine and valine, or corresponding keto acids (557, 10975 1112).
Affects dihydroxy acid dehydratase (22, 727) (Fig. 15). Most
alleles are leaky (557).
Leucine has a sparing effect on valine requirement (99). Called iv-1; groups 2
and 3.
ilv-2: isoleucine plus valine-2
VR. Closely linked to the right of ilv-1 (557). (482)
Requires both isoleucine and valine. Affects (alpha-keto-beta-hydroxylacyl
reductoisomerase (1112) (Fig. 15). Known alleles are not leaky (557). Allele T313 is
heat sensitive (557). Called iv-2; group 1.
ilv-3: isoleucine plus valine-3
IVR. Linked to met-2 (0/129) (PB). Between leu-2 (4%) and
ad-6 (9%) (579).
Requires both isoleucine and valine. Accumulates pyruvate. Very low acetohydroxy
acid synthetase activity (136) (Fig. 15). Alleles recombine and
complement (579, 1112).
Markedly inhibited by methionine (PB). Called iv-3; group 4.
In( ): inversion
Inversions can be used for mapping genes by duplication coverage (analogous to
deletion mapping). They are listed here only if they have contributed critical
information on gene sequences or locations of tips or centromeres. An inversion that
includes the centromere (i.e., that is pericentric) and has one breakpoint at a tip is
equivalent to a translocation in which a distal segment of one arm is transferred to the
tip of the other arm. Such an inversion of linkage group I is symbolized In(IL->
IR) or
In(IR-> IL). When such an inversion is crossed by normal sequence,
recombination
produces meiotic products that are duplicated for the transferred segment. Crosses
between two overlapping inversions produce recombinant meiotic products that are
duplicated for segments between the displaced breakpoints. For theory, diagrams,
and methods, see reference 808.
In(NM176): terminal pericentric inversion
In(IL->IR)NM176
A distal segment of IL is interchanged with the IR tip. Viable duplication progeny
from In x normal sequence contain two copies of the segment, which includes cys-5,
ser-3, and markers distal to them. but does not include un-3 or mt.
When In(NMI76) is
crossed with overlapping inversion In(OY323), an additional class of
duplication progeny
are produced that contain two copies of the intervals between the breakpoints of the two
inversions; see In(OY323) (57, 808, 1093).
In(OY323): pericentric inversion In(IL;IR)OY323
A long segment of I is inverted, which includes the centromere. When
In(OY323) is
crossed with overlapping inversion In(NMI76), viable duplication progeny are
produced
that contain two copies of the intervals between the breakpoints of the two inversions.
This duplicated segrnent includes ace-3 and nic-1 but not
lys-3 in IR, and leu-3 but not
nit-2 in IL. (57)
In(H4250): terminal pericentric inversion
In(IL->IR)H4250
A distal segment of IL is interchanged with the IR tip. Viable duplication progeny
from In x normal sequence contain two copies of the segment, which includes
suc and
markers distal to it but does not include phe-1 (761, 808).
inl: inositol
VR. Between pho-3 (3 to 4%) and pab-1 (1 to 10%). Right of
al-3 (362, 397, 1036).
(482)
Requires inositol (65). Lacks D-myoinositol-1-phosphatase (1142). Lack of
glucocycloaldolase found by Pina and Tatum (826) is attributed by Williams (1142) to
drastic repression of glucocycloaldolase by the concentration of inositol used for growth.
Growth is colonial on low levels of inositol (367). Tends to extrude dark pigment into
the medium when grown on suboptimal inositol. Composition of phospholipids
and cell walls is abnormal on limiting inositol (367, 439, 440, 501). Inhibited by
hexachlorocyclohexane (366, 457, 931). Conidia are subject to death by unbalanced
growth on minimal medium (1028, 1033), a property exploited for mutant enrichment
("inositol-less death") (606, 647) because double mutants are at a selective advantage.
Heat-sensitive allele 83201 is especially useful for mutant enrichment (832, 1043).
Used in the first experiments reporting transformation of Neurospora by
N. crassa DNA
(677, 679) and reported to be efficient as a recipient in absence of inositol (1162). Used
to study glucose (917) and sulfate (641) transport systems. Used extensively for studying
induced reversion (392). Used for studying the mechanism of inositol-less death (647,
702), mutagenicity of ferrous ions, and regulation of mitochondrial membrane fluidity;
for a review, see reference 702. Spontaneous reversion rates (386). Allele-specific
partial suppressor (390). Allele 46802 is nonrevertable and inseparable from
translocation 46802 (386, 808). Strains carrying heat-sensitive allele 83201 show slow
semicolonial growth in liquid minimal medium at 25°C (641), but look normal on
slants
(D.D. Perkins, unpublished data). Strains carrying allele 89601 contain cross-reacting
material (1183). Mutant gene exo-1 is present in the inl (89601) a
stock
FGSC 498 and
may, therefore, be present in stocks of mutants derived by inositol-less death. (See
references 194, 325, and 1027). Called inos.
inos: inositol
Changed to inl.
int: intense
IVR. Linked to pan-1 (0/50) (816).
Brighter orange than the wild type, perhaps because of morphology rather than
carotenoid content (816).
inv: invertase
VR. Right of pab-2 (3%) and ro-4 (5 to 8%), Left of
asn (4 to 9%) (918, PB).
Unable to use sucrose as a carbon source. Grows well on glucose or fructose and
fairly well on Casamino Acids or yeast extract. Invertase structural gene; invertase
deficient and uninducible by normal inducers. Makes cross-reacting material (919).
Invertase is also affected by cot-2, q.v.
ipa: "it pokes along"
IL. Between mt (20%) and arg-1 (1%) (994; E.G.
Barry, personal communication).
Hyphae from germinating ascospores or conidia grow for long distances without
branching. Cultures thus are 1 day late growing up (E.G. Barry, personal
communication). Modifies pro-3. Double mutant pro-3;ipa does not
respond to arginine
and grows less well than the single mutant pro-3;ipa+ on proline, citrulline, or
ornithine.
The single mutant ipa grows on minimal medium at half the wild-type rate.
Arginine
uptake is normal: arg-2;ipa or arg-5;ipa double mutants can grow on
arginine. Inhibition
studies suggest that ipa may be unable to shunt exogenous arginine into the
proline
pathway (994).
ipm-1: isopropylmalate permeation-1
Unmapped. Unlinked to ipm-2 or leu-4.
Able to use alpha-isopropylmalate to support growth of leu-4 mutants and
for
induction of alpha-isopropylmalate isomerase and beta-isopropylmalate dehydrogenase,
in contrast to ipm+ strains, which are unable to take up this intermediate
(870, 871).
ipm-2: isopropyimalate permeation-2
Unmapped. Unlinked to ipm-1 or leu-4.
Improves alpha-isopropylmalate uptake by the mutant ipm-1 in supporting
growth of
the mutant leu-4 (870, 871). By itself ipm-2 is not very effective in
promoting
permeability of alpha-isopropylmalate.
iv: isoleucine plus valine
Changed to ilv.
kyn-1: kynureninase
VII. Linked to nic-3 (30%) and wc-1 (20%) (G. Lester and P.J.
Russell, personal
communication).
Partially defective in induction of the kynurenase I (inducible) isozyme by kynurenine,
indole, or tryptophan, but has normal levels of the constitutive kynureninase II isozyme.
Possibly regulatory. Scored and selected by a low level of anthranilate accumulation on
medium supplemented with a high level of tryptophan, which results in a low
level of UV fluorescence compared with that of the wild type (926).
lac: lactose nonutilization
Multigenic basis. No major gene identified. Strains designated lac showed poor
growth on lactose. Lactase (beta-D-galactosidase) properties were unaltered; levels were
normal when grown on sucrose and depressed on lactose (584, 585). These strains differ
from wild type at several loci, each with a small and additive effect on lactose
utilization; e.g., three component genes from "lac-" strain 31389 x wild type
were shown
to be unlinked and were designated n-lac-1, pow(n-lac-2) (powdery
conidia), and
floc(nlac-3) (flocculent morphology). These genes are not specific for lactose
utilization
but also result in an altered adaptation response to other carbon sources and in other
pleiotropic effects. Thus, probably no identified locus qualifies to be designated
lac.
(357) The failure to find a single gene mutant unable to use lactose is ascribed to the
fact that Neurospora has two beta-galactosidases (605 and references
therein).
le-1: lethal ascospore-1
IVR. Linked to pan-1 (1 to 2%) (382). Right of cot-1 (14%)
(713, 716).
Autonomous ascospore lethal with colonial growth. Ascospores are black but mostly
fail to germinate. A few do so after special treatment (382). Aconidial colonial growth
with dense granular aerial mycelium, turning brown with age (713, 716). Photographs
(382, 716). Reduced amount of cell wall peptides (1165). Alleles B55 and S4355
of reference 382 were presumed to be allelic with similar mutations called
col-le-1
(CM3) and (col-le-2) by (713, 716), but direct tests were not made.
le-2: lethal ascospore-2
VIIL. Linked to met-7 (7%). Indicated left (382).
Autonomous ascospore lethal with colonial growth. Ascospores are black but mostly
fail to germinate, although a few are recovered after aging. Compact colonial growth
(382).
leu: leucine
For biosynthetic pathway, see Fig. 15. Leucine mutants have
been used extensively for
studies of regulation (see references 427 and 833). Leucine mutants acquire suppressors
when grown on Difco agar-sorbose medium. The suppressors are leaky auxotrophs
blocked at various steps in sulfur metabolism; apparently these blocks allow more
efficient use of the traces of leucine in the agar (425; S. R. Gross, personal
communication). Most aliphatic and aromatic amino acids can inhibit growth of leucine
mutants at appropriate concentrations, probably because of competition for a common
uptake system (S. R. Gross, personal communication). For regulation, see individual
loci; reviewed in references 427 and 642.
leu-1: leucine-1
IIIR. Between ad-4 (1 to 5%) and his-7 (8%) (219, 578, 815,
1052). (868)
Requires leucine (867, 868). Lacks beta-isopropylmalate dehydrogenase (426) (Fig. 15).
Accumulates alpha-isopropylmalate and beta-isopropylmalate (427). Synthesis of the
enzyme also requires the function of regulatory gene leu-3+ and the presence
of
alpha-isopropylmalate, which acts as inducer (427). Resistant to aminotriazole (D.
D. Perkins, unpublished data). Female sterile (O. M. Mylyk, personal communication).
Used to study reversion and competition in heterokaryons (901).
leu-2: leucine-2
IVR. Between trp-4 (2%) and ilv-3 (4%) (579, 991). (D.C.
Regnery, cited in
reference 633)
Requires leucine (867, 868). Structural gene for isopropylmalate isomerase (432, 871)
(Fig. 15). Altered heat inactivation of hybrid enzymes (432).
Structural differences
of hybrid enzymes (871). Accumulates alpha-isopropylmalate (427). Synthesis of the
enzyme also requires the function of regulatory gene leu-3+ and the presence
of alpha-isopropylmalate, which acts as an inducer (427). Resistant to aminotriazole
(D.D.
Perkins, unpublished data). Alleles show intralocus complementation (424). Allele
37501 is heat sensitive (30°C versus 20 C) and is leaky at 25°C (D.C.
Regnery, personal
communication).
leu-3: leucine-3
IL. Right of the In(OY323) left breakpoint and nit-2 (12 to 18%).
Left of cyt-1 (5 to
8%) and T(OY321) (57, 816, PB; D.D. Perkins, N.B. Raju, and E.G. Barry, in
preparation). (868)
Requires leucine (867, 868). Regulatory mutation; prevents synthesis of
alpha-isopropylmalate isomerase and beta-isopropylmalate dehydrogenase
and prevents full derepression of alpha-isopropylmalate synthetase; also involved in
regulation of isoleucine and valine synthesis, q.v. (427, 771,
833) (Fig. 15). The original allele, 47313, is leaky, but some
other alleles, e.g., R156, are
not.
leu-4: leucine-4
IL. Right of T(OY321); hence, of cyt-1. Left of cys-5
(<<1%) (1125; D.D. Perkins,
N.B. Raju, and E.G. Barry, in preparation). (429)
Requires leucine, Structural gene for alpha-isopropylmalate synthetase (426, 427, 432)
(Fig. 15). Feedback-negative mutants (426, 427). Hybrid
synthetases with altered properties
(432). Complementation between alleles (432, 1125).
leu-5: leucine-5
VR, Between cyh-2 (1%) and sp (3 to 9%) (818, 839, PB).
(812)
Strains carrying the only auxotrophic allele, 45208t. have a partial leucine requirement
at low temperatures and a tighter leucine requirement at 34°C and stop growth at
37 to
39 C, regardless of leucine supplementation (290, 839). Altered leucyl-tRNA synthetase
(839). Apparently a gene complex consisting of structural genes for cytoplasmic
leucyl-tRNA synthetase and a separate mitochondrial leucyl-tRNA synthetase.
Mutations mapping in the leu-5 region can affect either enzyme separately or
both
simultaneously (69, 431). In the [cni-3] mitochondrial mutant, mitochondrial
tRNA
synthetase is greatly increased, whereas the cytoplasmic enzyme is unchanged (430).
Allele 45208t causes alterations in unrelated enzymes, apparently via mistranslation (607,
839). Used to study the hypothesis that senescence is due to faulty protein synthesis
(607). Assembly of glycerolkinase and glycerol-3-phosphate dehydrogenase into inner
mitochondrial membrane is not impaired in leu-5 strains (213). Poor recovery
from
ascospores at 34°C (complex complete medium); best germinated at 25°C.
Allele 45208t
is somewhat unstable (431).
lis-1: light insensitive-1
IR. Between ad-3 (6%) and al-1 (16%) (775a; J. Paietta,
personal communication).
Circadian conidiation is not suppressed in constant light. Photoinduced
carotenogenesis and phase shifting of periodic conidiation are not altered. Recessive in
heterokaryons. (755a)
lis-2: light insensitive-2
VI. Between chol-2 (11%) and trp-2 (25%) (775a; J. Paietta,
personal communication).
Resembles lis-1.
lis-3: light insensitive-3
VR. Right of inl (4%) (775a; J. Paietta, personal communication).
Resembles lis-1.
lp: lump
II. Right of thr-3 (10%). Linked to bal (25%) (818,
812).
Restricted colonial growth. Differs from bal: faster growth, aerial hyphae
(812).
lys: lysine
All lysine auxotrophs are inhibited competitively by arginine (287, 288). Resistance to
arginine is conferred on lys-1 mutants by a presumed transport mutation
argR, q.v. See
the arg entry for medium that provides both lysine and arginine requirements.
Lysine
biosynthesis is by the alpha-aminoadipate pathway in Neurospora and other
higher
fungi (see reference 1103) (Fig. 16). Complex interactions
between lys, pyr,
and arg
mutations have been described (485). Enzymes of lysine biosynthesis are derepressed
coordinately with those of arginine, histidine, and tryptophan (1131). See
cpc-1.
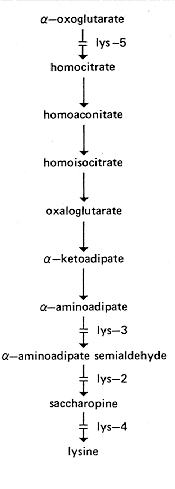
FIG. 16. Biosynthetic pathway of lysine, showing sites of gene action of
lys-4
and lys-5
and probable sites of lys-2 and lys-3 gene action (119, 400, 464, 762,
1087).
alpha-amino-epsilon-hydroxycaproic acid can be converted to alpha-aminoadipate
semialdehyde
(1178), but apparently is not an intermediate.
lys-1: lysine-1
V. Right of caf-1 (4 to 14%). Left of cyt-9 (5%) and
at (1 to 20%) (817; K.S. Hsu,
personal communication; PB). (403)
Uses lysine, alpha-aminoadipic acid, or epsilon-hydroxynorleucine
(alpha-amino-epsilon-hydroxycaproic
acid) (399, 400, 684, 1087). Accumulates homocitrate on limiting lysine concentrations
(464) (Fig. 16). Fine structure and complementation between
alleles (8). Initial allele:
33933.
lys-2: lysine-2
VR. Right of ilv-1 (4 to 7%). Left of cyh-2 (<1%) and
leu-5 (9%) (3, 818, 839). (399)
Requires lysine. Will not use epsilon-hydroxynorleucine (400). Probably blocked in
conversion of alpha-aminoadipate semialdehyde to saccharopine (1087) (Fig. 16). Initial
allele: 37101.
lys-3: lysine-3
IR. Right of al-1 (9%) and al-2 (12 to 15%). Left of
In(OY323) and nic-1 (<1%) (2,
907). Not included in duplications from In(OY323) x In(NM176);
hence, left of ace-3
(57). (288)
Uses lysine or epsilon-hydroxynorleucine. Probably blocked in conversion of
alpha-aminoadipate to alpha-aminoadipate semialdehyde, based on precursor utilization
(400,
1087) (Fig. 16). Complementation between alleles (13).
Ascospores are white and
inviable in homozygous lys-3 x lys-3 crosses, but some heteroallelic crosses are
fertile
(13). Inhibited by methionine. Initial allele: 4545.
lys-4: lysine-4
IR. Between nuc-1 (1%) and his-3 (1%) (271, 670).
(288)
Requires lysine (400). Lacks saccharopine-cleaving enzyme activity (1087) (Fig. 16).
Cornplementation between alleles (13). Use 0.5 mg of lysine per ml. Initial allele:
15069.
lys-5: lysine-5
VIL. Right of cyt-2 (6%), aro-6 (3%), and cpl-1 (5 to
7%). Left of un-4 (2%) (1012,
PB).
Requires lysine. Partial response to glutaric acid (762). Lacks homocitrate synthase
activity (464, 762) (Fig. 16). Some alleles such as 37402, called
asco, are
autonomous
ascospore lethals or semilethals, resulting in mostly immature white spores (see reference
1012 for a photograph). Viability of the lys- spores is improved by long
incubation (7).
With other alleles such as DS6-85, ascospores blacken and germinate normally.
Accumulates malate plus citrate on media with limiting lysine concentrations (464).
Four complementation groups (12). Allele 37402 called asco.
lysR: lysine resistant
IR. Between his-3 and nic-2 (C.C. Ho, personal communication).
(566)
Growth of the double mutant arg-1 lysR is resistant to the normal
inhibition by
L-lysine. Proposed to be due to basic amino acid transport system (566).
Possibly allelic with su(mtr)-1 (565).
Return to the FGSC Home page

Contact the FGSC
Last modified 4/24/96 KMC



