Neurospora mutants beginning with M through O
You may jump directly to the following sections:
Meiotic (mei)
Methionine (met)
Morphological (mo)
Methyl tryptophan resistant
Mutagen sensitive (mus)
Nicotinic acid (nic)
Nitrate non-utilization (nit)
Nuclease (nuc)
Osmotic (os)
m: microconidial
Allelic with pe, q.v.
ma-1: malate utilization-1
Unmapped. Probably in a left arm, any of III to VII.
Unable to use malate as a carbon source when the tricarboxylic acid cycle is blocked
by a suc (pyruvate carboxylase) mutation. Altered mitochondrial malate
dehydrogenase.
Scorable only in suc mutants (703, 706, 709).
ma-2: malate utilization-2
Unmapped. Unlinked to ma-1. Probably in a left arm, any of III to
VII.
Unable to use malate as a carbon source when the tricarboxylic acid cycle is blocked
by a suc (pyruvate carboxylase) mutation. Altered mitochondrial malate
dehydrogenase.
Scorable only in suc mutants (703, 706, 709).
mac: methionine-adenine-cystine
Probably allelic with met-6, q.v., but there are some differences between
them.
Requires methionine. Growth on methionine is stimulated by adenine with possible
additional stimulation by cystine. Inhibited by guanine (290). One occurrence: allele
65108.
mat: mat
IVR. Right of pyr-2 (3%). Left of the T(NMI52) right
breakpoint and cys-4 (10%)
(633, 721,812).
Spreading colonial morphology, with conidiating tufts. Grows better on sucrose
minimal medium than on glycerol complete medium (789).
Mating type
See A/a.
mb-1: male barren-1
VII. Linked to nic-3 and wc-1 (23%) (1100, PB).
Perithecial development is blocked when the mb-1 mutant is used as the
male parent
to fertilize an mb+ female: many perithecia are produced, mostly small,
brown, without
beaks or ascospores; a few perithecia mature and produce ascospores (1100). Perithecia
are reported to be normal and fertile when the mb-1 mutant is the female
parent
fertilized by an mb+ strain (1128); however, some abnormality as the female
has been
observed cytologically after pachytene (N.B. Raju, personal communication).
Homozygous barren (PB). Recessive in heterokaryons, complementing mb-2
and mb-3
(1101). One occurrence: allele 8455.
mb-2: male barren-2
IR. Between cyh-1 (5%) and al-1 (7%) (PB). (1100)
Perithecial development is blocked when the mb-2 mutant is used as the
male parent to
fertilize an mb+ female; many perithecia are produced, mostly small, brown,
without beaks
or ascospores; a few perithecia mature and produce ascospores (1100). Perithecia are
normal and fully fertile when the mb-2 mutant is used as the female parent
and fertilized
by an mb+ strain (1128). Homozygous barren (PB). Recessive in
heterokaryons,
complementing mb-1 and mb-3 (1101). One occurrence: allele
8553.
mb-3: male barren-3
IR. Linked to cyh-1 (18%), al-1 (2%), and mb-2 (6%)
(1100, PB; J.F. Leslie, personal
communication).
Perithecial development is blocked when an mb-2 mutant is used as the
male parent to
fertilize an mb+ female; many perithecia are produced, mostly small, brown,
without beaks
or ascospores; a few perithecia mature and produce ascospores (1100). Perithecia are
normal and fully fertile when an mb-2 mutant is used as the female parent
and fertilized by
an mb+ strain (1128). Development of perithecia may be slower than normal,
however,
when an mb-2 mutant is used as the female (N.B. Raju, personal
communication).
Homozygous barren (PB). Recessive in heterokaryons, complementing mb-1
and mb-3
(1101). Six occurrences.
mbic
See Bml.
md: mad
VR. Between sh (3%) and sp (9%) (296).
Spreading morphological which modifies the banding phenotype of cl.
Characteristic
branching pattern. Photographs. (296)
me: methionine
Changed to met.
mea-1: methylammonium resistant
Unmapped.
Presumed defective in transport of ammonium (293). nit-2;mea-1 double
mutants show
nitrogen-starved growth on ammonium (R.H. Garrett, personal communication).
med. medusa
IVR. Linked to met-5 (5%) and pan-1 (8%) (382).
Slow growing, spreading, morphological mutant, with distinctive grooves on the agar
surface (382). For a photograph, see Fig. 17 of reference 382. Reduced amount of cell
wall peptides (1165).
mei: meiotic
Meiosis impaired. In addition to partial or complete blocks of meiosis and ascus
development, some mutations designated mei have effects that may include sensitivity to
radiation, to radiomimetic drugs, and to histidine and increased duplication instability.
Both
recessive and dominant meiotic mutations are known. Mutations that affect meiotic or
premeiotic events may have other names, depending on the phenotype first recognized.
See:
asc; fmf-1; mb; mus-7, -8, -9, and -11; uvs-3, -5, and -6.
Meiotic mutants are very common
in natural populations of N. crassa (601).
mei-1: meiotic-1
IVR. Linked to arg-2 (<1%), probably to the right (995).
Meiosis is impaired in homozygous crosses. Recessive. Meiotic divisions occur and
many
ascospores are produced, but 70 to 90% are inviable and white. The viable ascospores
are
usually disomic for one or more linkage groups, indicating high nondisjunction at the first
division (254, 995). Chromosome pairing is defective: axial elements of synaptonemal
complex are present, but a completed complex is rarely seen. Separation at anaphases I
and
II is defective, leading to four-poled second and third division spindles (625). Not
sensitive
to UV, methyl methane sulfonate, ionizing radiation (939), or histidine (939; D.
Newmeyer,
unpublished data). mei-1 is present in wild-collected strain Abbott 4A (995),
which is an
ancestor of many Beadle and Tatum mutants (68). Possible allele asc(DL243)
complements
mei-1 but did not recombine with it (0/3,000) (253). DL243 and
mei-1 strains differ
phenotypically. In DL243 mutants, the major block is before karyogamy; the
few asci
produced have normal meiosis I but high nondisjunction at meiosis II, with most
chromosomes usually attached to only one spindle-pole body (254). Possible allele
asc(DL95) complements mei-1 and asc(DL243), but did
not
recombine with DL243 (0/96)
(253). DL95 is phenotypically like mei-1 but is less extreme (254).
Mei-2: Meiotic-2
VR. Linked near inl (995). Between al-3 (20 and his-6
(A.L. Schroeder, personal
communication).
Meiotic divisions occur, and many ascospor are produced, but many are inviable and
white.
Crosses heterozygous or homozygous for Mei-2 give extensive nondisjunction
of all linkage
groups (995). Chromosome pairing much reduced (B.C. Lu, cited in reference 995).
Sensitive to methyl methane sulfonate, histidine, and gamma rays (939). Dominant in
the
original strain (995), but progeny show incomplete penetrance (939).
mei-3: meiotic-3
IL. Right of arg-1 (3%). Probably right of eth-1 (1%) and
arg-3 (1%). Left of the T(39311)
right breakpoint; hence, left of the centromere and sn (757, 808).
Homozygous barren (757). Recessive. Blocks meiosis in zygotene (860). Sensitive to
UV,
histidine (757, 759), mitomycin C (195), ionizing radiation, and methyl methane sulfonate
(939). Sensitivity to UV and histidine is temperature sensitive; best scored at 39 C, at
least
for strains carrying allele N289 (757). Causes increased duplication instability (mitotic
recombination, deletion, or both) (757).
mei-4: meiotic-4
IIIR. Linked to leu-1 (12%), probably to left (757).
Homozygous barren. Recessive. Highly variable expression, depending on genetic
background (757). Crosses with more extreme genotypes are blocked at crozier
formation
or karyogamy (860). Crosses with less extreme genotypes complete normal first division
but
become irregular at later divisions, producing abnormal spores (B.C. Lu and D.R.
Galeazzi,
cited in reference 860). Not sensitive to UV, methyl methane sulfonate, or gamma rays
in
spot tests, or to histidine (757, 939).
mel-1: melon-1
VIIL. Left of thi-3 (27%) (819).
Growth as hemispherical colonies, similar to those of the mutant bal (717).
Growth
stimulated rather than depressed by sorbose (715). Cell wall analysis (278).
Photographs
(278, 717). Called col(C-L2b). Not tested by crossing to possible allele
do.
mel-2: melon-2
Allelic with bal, q.v. (812).
mel-3: melon-3
Linked to leu-1 (17%) (717).
Growth as hemispherical colonies similar to those of the mutant bal.
Photographs. A
separate modifier gene is also located in linkage group III. (717)
Mepr: methylpurine resistant
See mep.
mep(3): methylpurine resistant
Not mapped. Segregates 1:1.
Resistant to 6-methylpurine. Adenine phosphoribosyltransferase activity near normal in
vitro. Uptake is normal. 6-Methylpurine prevents purple pigment production by
ad-3 single
mutants on low adenine concentrations, but it does not prevent pigment production by
the
ad3A;mep(3) double mutant, suggesting that mep(3) results in an
alteration in the regulation
of de novo purine synthesis. Has low hypoxanthine phosphoribosyltransferase activity, as
do mep(10) and ad-8 mutants, q.v. Selected on 1 mM
6-methylpurine-sorbose
medium (785). Phenotype consistent with lowered affinity of glutamine amidotransferase
for 6-methylpurine as a feedback inhibitor (788). Not tested for allelism with
mep(10) or
with ad mutations. Called Mepr-3, but not indicated to be
dominant (785). Called mep-1
by Barratt and Ogata (51).
mep(10): methylpurine resistant
Not mapped. Segregates 1:1.
Resistant to 6-methylpurine. Adenine phosphoribosyltransferase activity is negligible
in
vitro. Adenine uptake is normal. Resistance may result from inability to convert the
analog
to nucleotide form. Has low hypoxanthine phosphoribosyltransferase activity, as do the
mutants mep(3) and ad-8, q.v. Selected on 1 mM
6-methylpurine-sorbose medium. Not
tested for allelism with mep(3). Called Mepr-10, but not indicated
to be dominant. (785)
Called mep-2 by Barratt and Ogata (51).
met: methionine
Auxotrophs designated met require methionine, and some can use its
immediate
precursors, homocysteine and cystathionine; they cannot use cysteine. (Mutants able to
use
cysteine as well as methionine are designated cys.) For the methionine
biosynthetic
pathway, see Fig. 17. For a review, see reference 351. For
regulation, see individual loci
and
reference 965. Formerly called me.
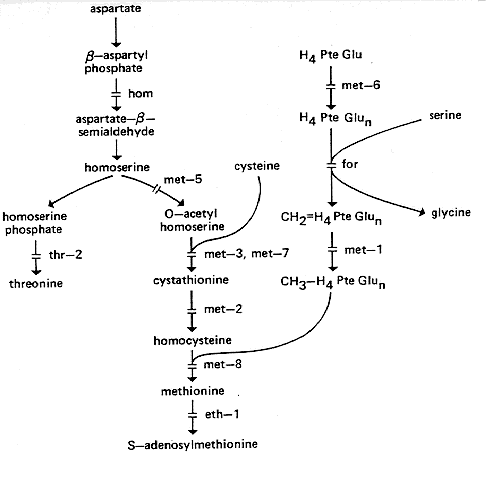
FIG. 17. Biosynthetic pathways of homoserine, threonine, and methionine,
showing sites
of
gene action (124, 208, 209, 351, 352, 518, 547, 965). For conversion of threonine to
isoleucine, see Fig. 15. H4PteGlu, Tetrahydrofolate. It is not clear whether the
polyglutamylation step controlled by met-6 occurs only at the stage shown.
met-1: methionine-1
IVR. Right of oxD (3%) and the T(S1229) left breakpoint. Left of
col-4 (4%) (55, 158,
718, 768, 808).
Uses methionine but not homocysteine (469) (Fig. 17). Lacks
methylene tetrahydrofolate
reductase and, thus, lacks the coenzyme needed for transmethylating homocysteine (124,
963, 964). A report that the mutant met-1 also lacks
cystathionine-gamma-synthase (547) proved
incorrect; the error resulted because methyl tetrahydrofolate is an essential activator of
cystathionine-gamma-synthase (965). Methylene tetrahydrofolate reductase is
feedback-inhibited by S-adenosylmethionine (124). Used in heteroallelic duplications
from
T(S1229) to assay mitotic recombination (56).
met-2: methionine-2
IVR. Between trp-4 (6%) and pan-1 (4%) (719). Linked to
ilv-3 (0/129) (354, 579).
Uses methionine or homocysteine; accumulates cystathionine (469). Lacks cystathionase
II
(353) (Fig. 17). Fine-structure map (720, 724).
Complementation map (719). Used in
major
studies of intralocus recombination and its polarity (720, 724).
met-3: methionine-3
VR. Right of trp-5 (4%) and pab-1 (1%). Left of pk
(1%) (6, 296, 362, 1036). (125)
Uses methionine, homocysteine, or cystathionine (469). Lacks
cystathionine-gamma-synthase
(547) (Fig. 17). This enzyme is also lacking in the mutant
met-7 (547). The
enzyme is
activated by methyl tetrahydrofolate and feedback inhibited by S-adenosylmethionine
(547,
965).
met-4: methionine-4
Changed to cys-10, q.v. (721).
met-5: methionine-5
IVR. Between his-4 (4%) and nit-3 (15%) (812, PB).
(125)
Uses cystathionine, homocysteine, or methionine (354, 718). Defective homoserine
transacetylase (547, 733) (Fig. 17).
met-6: methionine-6
IR. Right of T(NM103), T(ALS182), and thi-1 (7 to 14%) (808,
1091, PB). Left of ad-9 (2
to 16%) (466, 723, 789). Adjoining or allelic with mac (722, 724).
(125)
Requires methionine; does not use precursors (718; N. H. Horowitz, cited in reference
1180). Strain(s) carrying allele 35809 lacks polyglutamate forms of folate and, thus,
apparently lacks the coenzyme needed for transmethylating homocysteine (208, 886, 963,
964) (Fig. 17). An incorrect report that the mutant
met-6 also lacks
cystathionine-gamma-synthase (547) proved to be due to the methyl tetrahydrofolate
being
removed during preparation of extracts; methyl tetrahydrofolate is an activator of
cystathionine-gamma-synthase (965). The relationships between met-6 (35809)
and its
probable alleles met (S2706) and mac (65108) are not clear.
met (S2706) and mac both
complement met-6 (35809), but do not complement each other, indicating that
at least met
(S2706) and mac are alleles. In a high-resolution recombination study
with flanking
markers, met-6 (35809) and met (S2706) behaved like alleles, but
mac behaved atypically,
although almost equally closely linked (724). The mac mutant is reported to
differ from the
others in causing an accessory requirement for adenine and possibly cystine (290),
whereas
met-6
(35809) and met (S2706) strains are stimulated by adenine only in a
CO2-enriched
atmosphere (G. Roberts, cited in reference 724). mac and met-6
(35809) strains evidently
lack different folylpolyglutamate synthetase activities (208, 886). Used to study polarity
in
intralocus recombination (722, 724). Polarity with respect to flanking markers is not
reversed when the met-6 region is inverted relative to the centromere (722).
met-7: methionine-7
VIIR. Right of qa-2 (<1%), ars (<1%), and the centromere
(one
second-division ascus in
several hundred). Left of met-9 (10[-4]) and wc-1 (1 to 4%) (146,
725; M.E. Case, personal
communication). (718; M.K. Allen, cited in references 718 and 789)
Uses cystathionine, homocysteine, or methionine (718; N. H. Horowitz, cited in reference
1180). Lacks cystathionine-gamma-synthase (547) (Fig. 17).
This enzyme is also lacking
in the mutant met-3 (547). See met-3 for regulation. Apparently
contiguous with
met-9
by coconversion. Flanking markers are recombined in most met-7+
met-9+ recombinants
(725).
Functionally distinct from the mutant met-9, which has active
cystathionine-gamma-synthase
(547) but cannot use homocysteine. No mutants lacking both functions have been
isolated.
Allele NM251 is suppressible by supersuppressor RN33 (same as ssu-1?) (725).
Allele K79
is inseparable from reciprocal translocation T(I;VII)K79 (808).
met-8: methionine-8
IIIR. Between ff-5 (1 to 4%) and ad-4 (4%) (219, 815,1052).
(718)
Uses methionine but not precursors (718). Lacks methyl tetrahydrofolate homocysteine
transmethylase (124, 964) (Fig. 17).
met-9: methionine-9
VIIR. Between met-7 (10[-4]) and wc-1 (1 to 2%) (725).
(815)
Requires methionine; cannot use precursors (290, 718, 725). Apparently contiguous with
met-7, q.v. (725). Functionally distinct from met-7. The mutant
met-9 retains the
met-7+
function, producing cystathionine-gamma-synthase (547). Allele NM43 is heat sensitive
(725).
met-10: methionine-10
IR. Near lys-4. Right of nuc-1, T(AR173), and
his-2. Left of his-3 and ad-3 (757, 808; R.L.
Metzenberg, personal communication). (P. Dodd, cited in reference 816)
Requires methionine. The only known allele is heat sensitive, with the requirement at
34
C, not at 25°C (816; P. Dodd, cited in reference 816); does not grow at
39°C even with
methionine (757).
meth: methionine
Changed to met.
methionine overproduction
See eth-1 (542).
[mi-1]: maternal inheritance-1 (synonym: [poky])
Mitochondrial mutant with slow growth and deficient cyanide-sensitive respiration (see
reference 394). See su([mi-1]).
Microconidiation genotypes
Microconidia, being uninucleate, are valuable for such applications as somatic analysis
and
mutagenesis. They are much less abundant in the wild type than are multinucleate
macroconidia, except under certain conditions (893). Several genotypes are known that
increase the production of microconidia, notably, pe and dn, but
these single-mutant strains
also continue to produce macroconidia. A few microconidia, and no macroconidia, are
produced by the single mutant strains fl and cpt, q.v.; large numbers
can be obtained from
fl strains under certain conditions (893). Large numbers of almost exclusively
uninucleate
microconidia can be obtained by using the double mutants pe fl or
dn;fl, in which fl blocks
macroconidiation and the pe or dn component promotes
microconidiation. dn;fl strains have
the advantage over pe fl strains of greater fertility in homozygous crosses
(806), but
microconidia from dn;fl strains are less viable (454). Cultures abundantly
producing only
microconidia appear grey-brown rather than orange. See fl, dn, and
pe. For colonial
microconidiating strains, see col-1, col-4. See references 415 and 416 for other
gene
interactions.
mig: migration of trehalase
IR. Between met-6 (7%) and al-2 (20%). Between tre
(<<1%) and ad-9 (1045, 1176).
Altered electrophoretic mobility of trehalase (1045). Putative trehalase structural gene
(1047). (However, see qualifications in reference 1176.) Polymorphic in laboratory stocks
of N. crassa and in wild isolates of N. intermedia (1176). The
adjoining mutant gene tre
results in production of a protein inhibitor of trehalase (1045).
mo: morphological
Name and symbol used by Garnjobst and Tatum (382) for a miscellaneous group of
mutants
having spreading growth on agar, sometimes with scanty or fine hyphae and reduced
conidiation. The symbol morph has also been used. Other categories of
morphological
mutants were designated col, spco, or smco. Other workers have
assigned descriptive names,
e.g., bal, fr, ro, and sc. See also moe. For reviews
covering morphological mutants and
morphogenesis, see references 112, 197, 642, 675, 942, 946, and 1088. Growth rates and
hyphal diameters of 18 morphological mutants are given in reference 197.
mo-1: morphological-1
I. Linked mating type (9%) (382).
Altered morphology. Slow growth from ascospores (382).
mo-2: morphological-2
VII. Linked to nt (29%) and for (16%) (382, PB).
Slow growing, poorly pigmented mycelium. No conidia. Poor recovery from ascospores
(PB).
mo-3: morphological-3
See sk.
mo-4: morphological-4
IIIR. Right of leu-1 (8%). Linked to pro-1 (10%) (382).
(F.J. Doe, personal
communication)
Altered morphology. Conidiates throughout slant. Complements col-14,
col-16, and spg
(382).
mo-5: morphological-5
I. Linked mating type (20%) (382).
Few conidia. May make exudate on slant (382).
mo(KH160)
See shg: shaggy.
mo(P1163)
See dr: drift.
mo(P2402t)
See un-20.
mod-5: modifier of permeability
VI. Linked to trp-2, near the centromere (3%) (909).
Improves growth of trp-1, trp-2, trp-3, trp-4, aro-1, tyr-1, tyr-3, pt, met-7, and
pyr-1 strains
on complex media. Increases sensitivity to 4-methyltryptophan and
p-fluorophenylalanine.
Recessive in heterokaryons. Attributed to permeability change that facilitates entry of
metabolites (53, 909). Scorable on slants of minimal medium plus
4-methyl-DL-tryptophan
(0.9 mg/ml, autoclaved; tests read at 7 days 34 C) (PB). Map location similar to that of
mts, but not tested for allelism. mts differs in not allowing the
mutant pyr-1 (H263) to grow
on complex media (160). (Locus symbols mod-1, -2, -3, and -4 have
not been used.)
mod(sc): modifier of scumbo
IV. Linked to pan-1 (17%) (497).
Restricts the growth of sc but not of four other morphological mutants
(cr-1, fr, bis, sp) or
of the wild type (497).
moe-1: morphological, environment sensitive-1
Probably allelic with sk, q.v. VII. Linked to nt (12 to 19%)
(382), probably
to the right (PB).
Morphology identical to that of sk mutants; linkage similar (PB). Morphology
reported influenced by temperature and medium: more spreading on minimal or
complete medium at 25 C; zoned growth at 34°C on complete (382).
Temperature effect
could
be due to scot, the presence of which in the strain of origin was not
recognized. Photographs
of strain R2408: Fig. 19 through 22 in reference 382. Reduced amount of cell wall
peptides (1165).
moe-2: morphological, environment sensitive-2
VI. Linked to trp-2 (14%), probably to the left (382).
Grows with concentric zones on minimal medium and as restricted colonies on
glycerol complete medium (34 C). Photographs of strain R2532: Fig. 23 and 24 in
reference 382. The scot mutation may have been present in the strain of
origin.
moe-3: morphological, environment sensitive-3
IV. Left of pan-1 (17 to 25%) and bd (18%) (929).
Blocks conidial germination at high temperature. Colonial at high temperature
if on dialysis tubing on agar surface, but fairly normal vegetative growth if submerged.
Strong circadian conidiation rhythm at low temperature (929). Effect on conidial
germination
(but not on vegetative growth) counteracted by high conidial concentration or C02 (190,
929). Histidine is stimulatory, but there is disagreement as to whether it affects
germination
or vegetative growth (929; G.W. Charlang, personal communication). Partially curable
by
siderophores (ferricrocin). Conidia rapidly lose siderophores on contact with aqueous
medium, even at permissive temperatures, suggesting an alteration in the plasma
membrane
attachment site (190). Called JS134-9.
morph: morphological
Symbol changed to mo.
mt: mating type
See A/a. Also used as a symbol for mtr.
mtr: methyltryptophan resistant
IVR. Between pdx-1 (2%) and col-4 (1%) (101, 1017).
Resistant to 4-methyltryptophan and p-fluorophenylalanine. pmn (=
Pm-N, pm n), selected by resistance to p-fluorophenylalanine, has been shown
to be alletic with mtr
(R. Sadler and S. Ogilvie-Villa, personal communication; see also reference 248).
Defective
in transport of neutral aliphatic and aromatic amino acids via amino acid transport
system
I (as defined in reference 777) (248, 602, 1017, 1152). Causes an alteration in surface
glycoproteins (1038). Used extensively for transport studies (247a, 1150 [review], 1152),
also
for studies of the mechanism of intralocus recombination (1021). Resistance is recessive
in
duplications from T(S1229) (PB). Recessive resistance used in a heterokaryon
test
system
for mutation studies (1020). Suppressors obtained and used for selecting other resistance
mutants (106, 107, 555, 1018). Allele 26 is a putative frameshift mutation reverted by
ICR170 (106, 107). mtr ascospores are slow to darken and mature; up to 50%
of the young
ascospores from heterozygous crosses are white (152, PB). With probable allele MN18,
ascospore viability is improved by the addition of peptone to the crossing medium when
the
male parent is added (152). mtr has been scored on media containing 10 or 70
µg of
filter-sterilized 4-methyltryptophan per ml or on 20 or 60 µg of
p-fluorophenylalanine per
ml (550, 1021, PB). Unlike 4-methyltryptophan, p-fluorophenylalanine is heat stable and
can
be added before autoclaving. Strains with mutations at the mtr locus may be
obtained by
selection for resistance to numerous agents or for defects in uptake ability. Thus, there
is
confusion in nomenclature. Genes originally designated neua, neur, neut, tru
(628) may be
mtr alleles. mtr was initially called mt (602).
mts: methyltryptophan sensitive
VIL. Right of ylo-1 (<1%) (152, 160).
Sensitive to analogs of all tested aromatic, neutral, and basic amino acids and
to analogs of purines and pyrimidines. Ten to 100 times more sensitive than the wild
type.
Not sensitive to cold, salt, or detergent. Resembles mod-5 in enabling the
mutant trp-3
(A78) to grow well on complex media, but differs in not doing so for the mutant
pyr-1
(H263). No allelism test with mod-5. Obtained by filtration enrichment in the
presence of
5-methyltryptophan (152, 160). Used for selection of mutants resistant to analogs:
5-methyltryptophan (152); 8-azaadenine (524). Could be useful where the wild type is
not sufficiently
sensitive to allow direct selection of resistant mutants. Conveniently scored on
p-fluorophenylalanine (2 µg/ml, solid medium, autoclaved in medium;
p-fluorophenylalanine is more
heat stable than is 5-methyltryptophan). Best tested with small inocula on slants (10 by
75
mm) and read after 3 days at 34°C (PB). Called 5mt.
multicent
Linkage tester strain containing mt, bal, acr-2, pdx-1, at, ylo-1, and
wc-1, which
are linked to centromeres of linkage groups I through VII, respectively (800).
Especially useful to establish linkages of translocations (808). Scoring of test
markers is somewhat more laborious than for alcoy, which may therefore be
preferred for locating point mutations.
mus: mutagen sensitive
Symbol adopted in 1980. Locus numbers begin with mus-7 to avoid confusion
with uvs-1 through -6 (537, 539). Previously named
mutagen-sensitive genes
bearing other symbols retain their original designations in the present
compilation. (See uvs-1 to -6, upr-1, Mei-2, mei-3, nuh-4, and
gs.) Several new
unmapped mus genes (255) are not listed separately. For properties of double
mutants, see reference 539.
mus-7: mutagen sensitive-7
IIR. Between arg-5 (8 to 12%) and nuc-2 (11%) (539).
Sensitive to X rays, methyl methane sulfonate, and nitrosoguanidine, but not
to UV. Extremely sensitive to histidine. Normal spontaneous, UV-induced,
and X-ray-induced mutation. Homozygous barren (537, 539). Not tested for
allelism with asc(DL879), which maps in same region and causes
nondisjunction when homozygous.
mus-8: mutagen sensitive-8
IV. Linked to pdx-1 (6%) and mtr (1%) (537, 539; E.
Kafer, unpublished data).
Sensitive to UV, X rays, methyl methane sulfonate, nitrosoguanidine, and
mitomycin C. Decreased spontaneous mutation (537). Homozygous barren
(539).
mus-9: mutagen sensitive-9
IR. Between cyh-1 (18%) and al-2 (6%) (537).
Sensitive to UV, X rays, methyl methane sulfonate, histidine, nitrosoguanidine,
and mitomycin C. High spontaneous mutation; little or no mutability by UV
or X rays. Homozygous sterile. Reduced conidial viability. (537, 539)
Defective in extracellular nuclease, giving reduced halos around colony on
DNA agar (537). Initially called uvs(FK104) in reference 538.
mus-10: mutagen sensitive-10
VIIR. Right of met-7 (7%) (539).
Moderately sensitive to UV and methyl methane sulfonate. Not sensitive to
nitrosoguanidine or mitomycin C. Slight or no sensitivity to X rays or histidine.
Homozygous fertile (although less so than the wild type). Normal spontaneous
and UV- and X-ray induced mutation. (537, 539)
mus-11: mutagen sensitive-11
VR. Linked to pab-2 (539), near his-6 (E. Kafer, personal
communication).
Extremely sensitive to methyl methane sulfonate and histidine; also sensitive
to X rays, nitrosoguanidine, and mitomycin C (< X rays). High spontaneous
mutation. Little or no mutability by UV or X rays. Homozygous barren.
Reduced conidial viability (537, 539). Not allelic with Mei-2 (939).
mus(SC3): mutagen sensitive
Perhaps VI, linked to lys-5.
Sensitive to methyl methane sulfonate but not to histidine. Slow growth. Both
the mycelium and conidia are sensitive. Very sensitive on methyl methane
sulfonate medium, but not after treatment of conidia with methyl methane
sulfonate. (255) Allelism with mus(SC10) has not been excluded.
mus(SC10): mutagen sensitive
Sensitivity cosegregates with a translocation involving linkage groups II, III, and
VI.
Sensitive to methyl methane sulfonate, UV, and X rays. Sensitive to histidine
at 37°C but not at 25°C. High spontaneous mutation. Female sterile.
Complements uvs-4 (in IIIR) (255; A.M. De Lange, personal communication).
Allelism with mus-7 (II) or uvs-5 (111) is not excluded.
mus(SC15): mutagen sensitive
V. Left of inl (10%) (255).
Highly sensitive to methyl methane sulfonate but not to histidine (255).
Sensitive to X rays (A. M. De Lange, personal communication). Both the
mycelium and conidia are sensitive. Very sensitive on methyl methane
sulfonate medium; the effect is slight after treatment of conidia with methyl
methane sulfonate (255). No allelism test with mus(SC17).
mus(SC17): mutagen sensitive
V. Left of inl (27%) (255).
Sensitive to methyl methane sulfonate but not to histidine. Sensitivity is shown
by the mycelium, not by conidia, and only after preincubation at 15°C. Growth
is cold sensitive on minimal medium (255).
mus(SC28): mutagen sensitive
IR. Right of al-1 (18%) (255).
Sensitive to methyl methane sulfonate. Both the mycelium and conidia are
sensitive (255).
nada: NAD(P)ase
IV. Left of ad-6 (18%) (747).
NAD(P) glycohydrolase structural gene. Normal morphology. Identified by
a plaque test, using Haemophilus influenzae. Recessive in heterokaryons.
Allele 62ts is temperature sensitive, with altered substrate affinity. (747) Used
in a study of glutamic acid decarboxylase during conidial germination (196).
nap: neutral and acidic amino acid permeability
VR. Linked to inl (15%) (516); right of ure-2
(32%) (1149).
Selected as resistant to ethionine plus p-fluorophenylalanine (516). Causes
reduced amino acid uptake by neutral, basic, and general systems. Also causes
reduced uptake of uridine and glucose. Defect is not in amino acid-binding
glycoproteins. (865) See reference 1149 for aspartate uptake and resistance to
inhibitors. Scored by spotting conidial suspension on minimal medium plus
1.5% sucrose, agar, 0.3 mM ethionine, and 0.02 mM p-fluorophenylalanine.
nd: natural death
IR. Between the centromere (15%) and al-2 (20%) (981).
Decreasing clonal growth potential under all nutritional conditions, followed
by abrupt irreversible cessation of growth (707, 981). Hypersensitive to
sorbose. Conidia die rapidly on slants at 4°C (707). Recessive in
heterokaryons. An aged strain can be rejuvenated through heterokaryosis or
by crossing to nd+. Extracts nontoxic (981). Used to examine
hypotheses of senescence based on faulty protein synthesis (607) and lipid
autoxidation with free-radical reactions (702). Stocks maintained in balanced
heterokaryons. Initial growth rate of the original strain, 2.5 mm/h; however,
nd progeny free of modifiers grow initially at 4.5 mm/h (wild-type
rate) (707).
ndc-1: nuclear division cycle-1
VR. Left of arg-4 (2%) (976).
Heat-sensitive conditional mutant. Growth at 25°C but not at 34°C.
Recessive.
Division
cycle blocked just before initiation of DNA synthesis while spindle-pole bodies
are
duplicated but not separated. (976) Scored as an irreparable un
mutant (see un).
neu: neutral amino acid transport
See mtr.
nic: nicotinic acid
nic mutants are preferably supplemented with nicotinamide rather
than nicotinic acid at
most pH values because of permeability (97). nt mutants are best
treated as nic mutants
for purposes of growth and scoring. For good recovery of some nic
mutants from crosses, crossing media should be supplemented with
nicotinamide at levels higher (10X) than those required for growth, even when
the protoperithecial parent is nic+ (789; P. St. Lawrence, personal
communication). See Fig. 18 for the biosynthetic pathway.
For regulation, see references 111, 371, 604,
and 926.
nic-1: nicotinic acid-1
IR. Right of ace-3 (<1%), lys-1 (1%), and
In(OY323). Left of os-1 (10 to 29%) (2, 57,
131, 578, 789, 816, 907). (482)
Uses nicotinic acid or nicotinamide, but not precursors (97, 100) (Fig.
18).
Accumulates quinolinic acid (100). Used to study intralocus recombination
(907).
Called the q locus.
nic-2: nicotinic acid-2
IR. Between ad-3B (4%) and ace-7 (4 to 7%) (271, 578).
(482)
Grows on nicotinic acid, nicotinamide, or high concentrations of quinolinic acid
(97,
1168). Cannot use kynurenine, hydroxykynurenine, or hydroxyanthranilic acid
(96, 1168).
Accumulates 3-hydroxyanthranilic acid (96) (Fig. 18). Aging
cultures
accumulate
red-brown pigment in the medium. Used to study intralocus recombination
(908).
Translocations T(4540) and T(S1325) are inseparable
from nic-2 (808, 908, 911).
nic-3: nicotinic acid-3
VIIL. Right of spco-4 (1%) and do (3%). Left of
thi-3 (9 to 27%) and csp-2 (16 to
22%) (539, 812, 816, PB). (M.K. Allen, cited in references 718 and
789)
Uses nicotinic acid, nicotinamide, 3-hydroxyanthranilic acid,
3-hydroxykynurenine, or high
concentrations of quinolinic acid (96, 1168). Accumulates
alpha-N-acetylkynurenine;
blocked in conversion of kynurenine to 3-hydroxykynurenine (1168) (Fig. 18).
Pyridine
nucleotide levels (111).
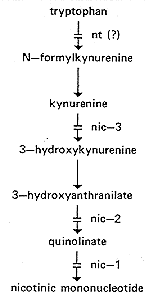
FIG. 18. Pathway from tryptophan to nicotinic mononucleotide,
showing sites of gene
action (96, 100, 368, 1168). The enzymatic reactions between
3-hydroxyanthranilate and
nicotinic mononucleotide have not been demonstrated directly in Neurospora.
nit: nitrate nonutilizer
Conveniently scored on synthetic crossing medium (1134), in which nitrate is
the sole
nitrogen source. Also scorable on slants by pH change when grown with
ammonium
nitrate as the nitrogen source and bromcresol purple (4 mg/ml) as an indicator
(791). In
most crosses, a nit mutant can be used as the fertilizing parent; in
nit x nit or other
crosses where it is required as the female parent, crossing medium can be
altered by
substituting ammonium nitrate for potassium nitrate (155). Nitrite is toxic at
low pH;
test media containing nitrite should be neutralized, and the nitrite should
preferably be
filter-sterilized (G.S. Sorger, personal communication). For a summary of
nutritional
requirements, based on the data of various authors, see reference 1080.
nit-1, nit-7,
nit-8, and nit-9 involve a molybdenum-containing cofactor
common to nitrate reductase
and xanthine dehydrogenase (591, 1080, 1081) (Fig. 19 and 24). For a review
of nitrate
assimilation, see reference 385. For regulation, see reference 643 (review),
references 292, 835, 837, and 1081, and entries for nit loci,
gln-1, and nmr.
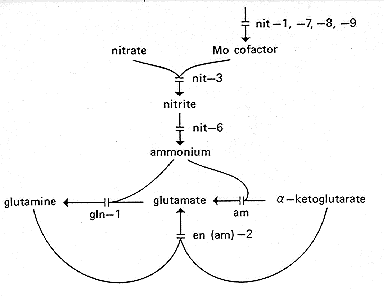
FIG. 19. Nitrate reduction pathway showing sites of gene action.
am only blocks the
NADP-specific glutamate dehydrogenase, not the NAD-specific enzyme. (185,
293, 336,
591, 912, 999, 1081)
nit-1: nitrate nonutilizer-1
IR. Right of Tp(T54M94) and ad-9 (3 to 15%). Left of
cyh-1 (6%) (466, 496, 816). (482)
Cannot use nitrate or hypoxanthine as a nitrogen source, but uses nitrite,
ammonia, or
amino acids (1000). Does not prevent formation or nitrate reductase
apoprotein (999),
but lacks the molybdenum-containing cofactor common to nitrate reductase
and xanthine dehydrogenase (591, 741) (Fig. 19 and 24). The nitrate reductase
in nit-1 extracts does not catalyze the complete electron transport
sequence from NADPH to N03 but does catalyze the initial part of this
sequence if a
suitable electron acceptor (e.g., cytochrome c) is provided (999). See reference
198 for a model of interaction of nit-1 and nit-3 gene
products. See references 226, 999, and 1000 for regulation.
nit-2: nitrate nonutilizer-2
IL. Right of the T(39311) left breakpoint and of un-5
(2%). Left of In(OY323) and
leu-3 (12 to 18%) (57, 808, 816, PB). (335, 1135)
Cannot use nitrate, nitrite, purines, or most amino acids as a nitrogen source
but will
grow on ammonia, glutamine, or glutamate. nit-2+ is a major
nitrogen control gene and
mediates nitrogen catabolite repression. The nit-2 mutant is missing
(or has severely reduced levels of) nitrate reductase, nitrite reductase, uricase,
xanthine dehydrogenase, allantoinase, allantoicase, L-amino acid oxidase,
general amino acid permease, extracellular protease, and an intracellular
neutral
phenylmethylsulfonyl fluoride-sensitive protease (227, 324, 441, 872, 1001, and
references therein). Also affects levels of glutamate dehydrogenases (226) and
uptake of uracil and uridine (128). Prevents leaky growth of the mutant
am on minimal medium (155). The product of the nit-2
gene has been
tentatively identified as a nuclear DNA-binding protein, whose affinity for
DNA is
reduced in the presence of glutamine (433). Allele K31 (called pink)
originated in N. sitophila and was introgressed into N.
crassa (335); protein product of K31 may show altered mobility (433).
Recombination within the nit-2 locus is subject to regulation by
rec-1 (157).
Heterozygosity for closely linked ss reduces recombination within
nit-2 (161). Called amr: ammonium regulation in
reference 872.
nit-3: nitrate nonutilizer-3
IVR. Between met-5 (15%) and pyr-2 (2 to 9%) (1000,
PB). (453)
Cannot use nitrate as a nitrogen source, but uses nitrite, ammonia,
hypoxanthine, or
amino acids (28, 999). Structural gene for NADPH nitrate reductase (28) (Fig. 19).
Allele 14789 apparently codes for an altered enzyme that cannot catalyze the
whole electron transport sequence from NADPH to N03-, but can catalyze the
terminal portion of this sequence, providing that a suitable electron donor
(reduced viologen dye) is provided (999). See reference 198 for a model of
interaction of nit-1 and nit-3 gene products. For
regulation, see references 226, 999, and
1000. The nit-3+ gene has been cloned and is expressed in
Escherichia coli (989).
nit-4: nitrate nonutilizer-4
IVR. Right of pyr-1 (1 to 6%). Probably right of col-4
(2%). Left of pan-1 (6 to 27%)
(1000, PB). (94)
Cannot use nitrate or nitrite as a nitrogen source, but uses ammonia and
amino acids
(94). Regulator for induction by nitrate of nitrate reductase and nitrite
reductase (1080).
Allele nr15, called nit-5 (1000), is phenotypically identical to other
nit-4 alleles; shown to
be allelic by failure to complement or recombine (0 prototrophs per 2,080
progeny)
(1080). Original allele was discovered in a wild isolate of N.
intermedia from Borneo
and introgressed into N. crassa (94).
nit-5
Allelic nit-4, q.v.
nit-6: nitrate nonutilizer-6
VIL. Right of chol-2 (6%). Left of ser-7 (10%) and ad-8
(17%) (PB; O.C.
Yoder, personal communication).
Unable to use nitrate or nitrite as a nitrogen source (185). Lacks nitrite
reductase (185) (Fig. 19), which is subject to positive nitrogen
metabolite
repression (186). Affected by nit-2 and MS5 regulator genes (838, 1076a)
(see nmr-1). Used to study repression of nitrate reductase (26) and
nonenzymatic reduction of nitrite (185). Induced by nitrite (198).
nit-7: nitrate nonutilizer-7
IIIR. Between trp-1 (26 to 32%) and dow (45%) (D.D. Perkins,
unpublished
data).
Cannot use nitrate or hypoxanthine as a nitrogen source. Resembles nit-1,
nit-8, and nit-9 in affecting the molybdenum-containing cofactor
common to
nitrate reductase and xanthine dehydrogenase (1080, 1081) (Fig.
19 and 24).
nit-8: nitrate nonutilizer-8
IR. Linked to nit-1 (32%) (1080). Right of mt (10 to
15%) (D.D. Perkins,
unpublished data).
Cannot use nitrate or hypoxanthine as a nitrogen source. Lacks the
molybdenum cofactor for nitrate reductase and xanthine dehydrogenase (1080,
1081) (Fig. 19 and 24).
nit-9: nitrate nonutilizer-9
IVR. Right of nit-4 (9%). Linked to nit-3 (35 to 38%)
(1080).
Cannot use nitrate or hypoxanthine as a nitrogen source. Lacks the
molybdenum cofactor for nitrate reductase and xanthine dehydrogenase (Fig. 19 and 24). A complex locus
with
three complementation groups, comparable
to cnxABC of Aspergillus nidulans. (1080, 1081)
nmr-1: nitrogen metabolite regulation
VR. Between am (3 to 7%) and gln-1 (4 to 10%) (1079).
Synthesis of nitrate reductase is derepressed on ammonium, glutamate, or
glutamine. Hypostatic to nit-2 and nit-4. Prototrophic. Isolated
and scored
by sensitivity to chlorate in the presence of glutamine. (295, 1079)
MS5 is a possible allele. MS5 is unmapped and is not allelic with nit-2 or
nit-3 on the basis of two wild-type ascospores from poorly fertile crosses in
each case. MS5 is derepressed on glutamine. Levels of nitrate reductase,
nitrite reductase, histidase, and acetamidase are elevated in the presence of
glutamine and the respective enzyme inducer. Prototrophic. Scored the same
as the mutant nmr-1 and checked by assaying for nonrepressibility of
nitrate reductase by glutamine. (838; G.J. Sorger, personal communication)
NO: Nucleolus organizer
VL. Right of terminal sat (60, 817) and of terminal translocations
ALS176,
ALS182, and AR190. Left of lys-1 and of translocations
AR30, AR33, AR45,
NM130, AR177, and NM183; thus, left of caf-1 (60, 817; D.D.
Perkins,
N.B. Raju, and E.G. Barry, in preparation). T(OY321) divides the NO into
two portions, each of which retains ability to form a nucleolus (D.D. Perkins,
N.B. Raju, and E.G. Barry, in preparation). (821)
Genes specifying 5.8S, 17S, and 26S rRNA (but not 5S) are located in the
nucleolus organizer region in a tandemly repeated DNA sequence (215, 361).
Wild type 74-OR23-1A has 185 tandem repeats (571). Nucleotide sequence
of the 5.8S ribosomal DNA has been determined; comparison with yeast cells
shows 145 of 158 rRNA residues conserved (974). Hybridization shows that
sequences are shared both with Xenopus and Drosophila (216).
T(AR33)
produces duplications with two copies of the nucleolus organizer (817), which
undergo demagnification (887) in such a way that different nontranscribed
spacer sequences from both parental nucleolus organizers are retained (888).
Genes specifying 5S rRNA are not included in the ribosomal DNA repeat unit,
but are located elsewhere in the genome as dispersed single genes surrounded
by heterogeneous flanking sequences (361, 975). For rRNA processing, see
rip-1.
nt: nicotinic acid or tryptophan
VIIR. Between arg-10 (2 to 12%) and sk (7 to 18%) (789).
(874)
Uses nicotinic acid. May respond also to tryptophan, phenylalanine, tyrosine,
quinic acid, and precursors of nicotinic acid or tryptophan, or both, depending
on genetic background (448, 760). Best supplemented with nicotinamide and
scored as a nic mutant. Probably deficient in tryptophan pyrrolase (tryptophan
2,3-dioxygenase) (Fig. 18), but direct evidence is lacking
because tryptophan
oxygenase cannot be assayed in Neurospora (368). Kynurenine formamidase
levels are normal (368). Pyridine nucleotide levels (111).
nuc-1: nuclease-1
IR. Right of T(AR173) and his-2 (<1%). Left of lys-4
(1%). (514)
nuc-1 mutants (other than nuc-1c) are unable to use RNA or DNA
as a
phosphorus source (450, 514). Defective in production of repressible alkaline
and acid phosphatases (671, 1077). Several nucleases are absent or reduced
(449). nuc-1 is epistatic to both pconc and pregc (671)
and to pgovc (665).
Scored on low-phosphate medium by a staining reaction with alpha-naphthyl
phosphate plus diazo blue B (397, 1077), by failure to grow on minimal
medium altered so that 0.1 g of RNA or DNA per liter is substituted for the
inorganic phosphate source (514, 538), or by failure to grow on low-phosphate
medium at a pH above 7 (R.L. Metzenberg, personal communication).
nuc-1c is constitutive for alkaline phosphatase synthesis and maps very close
to nuc-1. nuc-1c acts only if it is cis to normal nuc-1. In
duplications, nuc-1c
is dominant to nuc-1+, which is dominant to nuc-1. nuc-1c is
epistatic to nuc-2
(670). nuc-1c is scored on high-phosphate medium by a staining reaction with
alpha-naphthyl phosphate plus diazo blue B (397, 1077) or by suppression of
the nuc-2 phenotype on low-phosphate medium at high pH (670). Used to
study phosphate transport (624). For regulation model see references 665 and
670.
nuc-2: nuclease-2
IIR. Between the T(NM177) breakpoints; hence, right of aro-3.
Left of preg
(1 to 2%) and pe (4%). Probably allelic with pcon (0/854) (593,
671). (514)
Unable to use RNA or DNA as a phosphorus source (514). Defective in
production of repressible alkaline and acid phosphatases (671, 1077). Several
nucleases absent or reduced (449). Interaction with other phosphate
regulatory genes (665). Recessive to nuc+ in partial diploids and
heterokaryons (671). Not defective in nuh function (538). Scored on
low-phosphate medium by a staining reaction with alpha-naphthyl
phosphate plus diazo blue B (397, 1077), by failure to grow on minimal
medium altered so that 0.1 g of RNA or DNA per liter is substituted for the
inorganic phosphate source (514, 538), or by failure to grow on low-phosphate
medium at a pH above 7 (R.L. Metzenberg, personal communication). Used
to study phosphate transport (624). For a regulation model, see references 665
and 670. See pcon.
nuh: nuclease halo
Deficient in extracellular nuclease, giving reduced halos around colonies on
DNA agar (538). The mutant nuh-4 is also sensitive to UV and
nitrosoguanidine; the others are not. However, two mutants isolated by UV
sensitivity, uvs-3 and uvs-6, also have the nuh phenotype
(538).
nuh-1: nuclease halo
IIIR. Right of leu-1 (4%). Left of nuh-2 (<1%) and
trp-1 (11%) (538).
nuh-2: nuclease halo-2
IIIR. Right of nuh-1 (<1%) and leu-1 (4%).
Left of trp-1 (11%) (538).
nuh-3: nuclease halo-3
VR. Between cyh-2 (4%) and al-3 (17%) (538).
Releases only small amounts of deoxyribonuclease A (endonuclease) and
deoxyribonuclease C (endo-, exonuclease) (359). Not sensitive to UV or
chemical mutagens (E. Kafer, cited in reference 195).
nuh-4: nuclease halo-4
Probably allelic with uvs-3, q.v. (537, 538).
nuh-5: nuclease halo-5
IIR. Linked to trp-3 (30%), near T(4637) (538).
nuh-6: nuclease halo-6
IR. Between the centromere (5%) and nic-2 (4%) (538).
nuh-8: nuclease halo-8
IR. Right of nic-1. (See note added in proof in reference 538).
Formerly called nuh(18).
nuh(23): nuclease halo
VR. Linked to nuh-3 (6%) (538).
Nystatin resistant
See erg: ergosterol.
oli: oligomycin resistant
VIIR. Between met-9 (8 to 24%) and arg-11 (3 to 16%) (960).
Linked to
frq-1 (<2%) and possibly allelic (282, 283). (959)
Resistant to oligomycin. Defective in energy transduction (313). Structural
gene for dicyclohexylcarbodiimide-binding proteolipid (subunit 9) of F0 portion
of mitochondrial adenosine triphosphate synthetase (958). The amino acid
sequence (81 residues) (959) shows extensive homology with the corresponding
proteolipid in yeast, in which, in contrast to Neurospora, it is the product of a
mitochondrial gene (960, 1109). Specific single amino acid substitutions have
been identified for three mutants (959). oli mutants are selected effectively by
using double mutant azs; has (called ANT-1), which is deficient in both salicyl
hydroxamic acid-sensitive and azide-sensitive alternate oxidase pathways.
Scored on 5 µg of oligomycin per ml of liquid medium, 3 days, 30°C (312).
Altered period of circadian rhythm cosegregates and reverts with oli (282,
283).
os: osmotic sensitive
Unable to grow on media with elevated osmotic pressure. Scorable on solid
or liquid media plus 4% NaCl (1.4 M). Most alleles can also be scored by
morphology, having sticky, close-cropped aerial hyphae that tend to rupture
and bleed. Morphology is influenced by humidity. Intense pigment of
aggregated hyphae has suggested the name "flame," which was originally
applied to some os mutants. os strains are useful for obtaining
protoplasts
(e.g.,
reference 971) and are reported to be efficient as recipients for DNA-mediated
transformation in media of high osmolarity (1162). In addition to the
numerous loci designated os, cut is a typical osmotic mutation. Mutant
sor(T9) is osmotic sensitive and sorbose resistant and has low glucoamylase
activity, but does not show the typical os morphology.
os-1: osmotic-1
IR. Between nic-1 (10 to 29%) and arg-13 (1%) (789, 812, 816).
(M.R.
Emerson, cited in reference 789)
Sensitive to high osmotic pressure. Readily scored by morphology on nonmoist
slants or by failure to grow on media with 4% NaCl. Most os-1 alleles result
in cultures that form no or few conidia on agar slants. Alleles
NM233t and NM204t are heat sensitive (25°C versus 34°C). In media of
high
osmolarity, os-1 strains form protoplasts (323, 438). os-1 (Bl35) is an
essential
genotypic component of the wall-less strain slime (321). Protoplasts of strains
carrying heatsensitive allele NM233t are stable at 37 C, with a 7.5-h redoubling
time, and show good regeneration. The biochemical defect differs from that
affected by either polyoxin or sorbose (chitin or glucan syntheses) (970, 971).
Cell wall pores are four times larger in an os-1 mutant than in the wild type;
os-1 also has a higher exclusion threshold and a 30-fold-higher galactosamine/
glucosamine ratio (1083, 1084). Intralocus complementation (676). Allele
Y256M209 called flm-1.
os-2: osmotic-2
IVR. Right of cot-1 (4%) (816; A.L. Schroeder, personal
communication).
Sensitive to high osmotic pressure. Readily scored by morphology on nonmoist
slants.
os-3: osmotic-3
Described as a IR mutation right of nic-2 (4%) (654). Because of stock loss
and ambiguity, validity as a separate locus cannot be confirmed (655, 802).
os-4: osmotic-4
I. Right of T(39311) and arg-1 (1%). Left of T(AR173)
and T(AR190); hence,
of un-2 and his-2. Linked to sn (0/33). (Data for allele
Y256M233.) (802, PB)
Sensitive to high osmotic pressure. Readily scored by morphology on nonmoist
slants. Allele Y256M223, originally called flm-2, is preferred over NM201o,
on which the locus designation was initially based. (802)
os-5: osmotic-5
IR. Right of cyh-1 (12%). Left of the Tp(T54M94) right breakpoint
and of
arg-6 (1%). Linked to al-2 (<1%). (802, 808)
Sensitive to high osmotic pressure. Scorable by morphology on nonmoist
slants.
os-6: osmotic-6; os-7: osmotic-7
Symbols used in reference 676 for os-1-linked osmotic mutations obtained
among inl+ transformants in experiments using wild-type
Neurospora DNA.
Osmotic and osmotic-like mutants have also been reported in other
transformation experiments (1162). It seems wise not to define new loci on the
basis of variants arising in transformation experiments or to use data from
transformed strains or their derivatives in mapping. Accordingly, os-6 and
os-7
have not been included in the list of established loci.
ota: ornithine transaminase
IIIR. Between ad-4 (15%) and tyr-1 (14%) (241). Linked to
pro-4 (4%) (D.J.
West, cited in Neurospora Newsl. 16:19-22, 1970).
Ornithine-delta-transaminase deficient (241) (Fig. 10).
Conidiates somewhat
less than does the wild type (S. Brody, personal communication). Selected by
ability to use exogenous ornithine as a precursor for arginine in an arg-5
arg-12s double mutant. Catabolism of ornithine (to
glutamic-gamma-semialdehyde) is blocked, resulting in ornithine
concentrations high enough to compensate for the low activity of the ornithine
carbamyl transferase in the arg-12s mutant. The ota single mutant
is
prototrophic but prevents the efficient use of ornithine or arginine as the sole
nitrogen source (241). Used to study flux through the arginine biosynthetic
pathway (401). Used to study the utilization of endogenous versus exogenous
ornithine (234). Sideramine production is completely blocked in absence of
ornithine in the ota;arg-5;aga triple mutant, which is used to study iron
transport (1146, 1147).
oxD: D-amino acid oxidase
IVR. Between the T(S1229) breakpoints; hence, right of pdx-1
(0/55 asci).
Left of met-1 (3%) (55, 768, 808).
Lacks D-amino acid oxidase. Unable to use D-methionine to satisfy the growth
requirement of the mutant met-1. Increased sensitivity to toxic effects Of
D-phenylalanine and D-tyrosine. Unable to use D-methionine as the sole
sulfur source (768). Resistant to D-ethionine (477). Strains carrying allele
oxD1 are cysteine auxotrophs, probably owing to a closely linked coincident
lesion (768); see cys-15.
Oxidase, terminal
See aod, azs, has, and cni-1.
Return to the FGSC Home page

Contact the FGSC
Last modified 4/24/96 KMC



