LINKAGE MAPS
Jump ahead to any of the linkage groups:
Linkage group I
Linkage group II
Linkage group III
Linkage group IV
Linkage group V
Linkage group VI
Linkage group VII
Genetic linkage information on chromosomal loci is summarized in
Fig. 1 through 7. Loci whose order is established with reasonable
certainty are displayed on the maps. Many loci are not displayed, but
are listed below the maps because their order has not been completely
established relative to loci shown on the map. The meaning of each
locus symbol, data on linkage with nearby markers, and further detailed
information will be found in the section Information on Individual Loci,
where genes are listed alphabetically by symbol.
Relative distances on the maps are only rough approximations. rec
genes with local effects differ in many of the strains used for mapping,
and recombination values for the same interval have been shown to vary
10-fold or more in crosses of different parentage (167, 169, 170, 174).
For this reason, no attempt has been made to correct for undetected
multiple crossovers in long intervals by using a mapping function.
Linkage group I is estimated to be at least 200 map units long, and
the total for all seven groups probably exceeds 1,000 (808). Because of
the genetically determined variability, no map scale is indicated
graphically. Interval lengths were estimated as composite or
representative values based on all the crosses available and are shown
on the scale ca. 1 cm = 7% recombination. It must be stressed that
absolute and relative interval lengths shown on the maps possess only
limited predictive value, depending on the genotype of a particular cross
with respect to genes that determine the frequency of recombination in
each local region. In contrast, gene order is constant in the absence of
rearrangement.
Because of rec gene differences, gene order cannot be established
reliably by combining two-point recombination values from different
crosses. Seriation has, therefore, been based wherever possible either
on meiotic crossing over data from three-point crosses or on duplication
coverage. Duplication-producing rearrangements such as insertional or
terminal translocations allow a simple test to be performed from which
it can be determined whether a locus is to the right or left of a
rearrangement breakpoint (see, for example, references 798, 808, and
816). Breakpoints are shown on the maps only for those
rearrangements that have been used to establish gene order or to map
chromosome tips and centromeres.
Crosses involving strains with a history of transformation have not
been used in constructing standard maps because genes may be inserted
in abnormal positions, and we recommend that new loci not be defined
on the basis of mutations that originated in pedigrees from transformed
parents until more is understood about the transformation process. (See
the entry for os-6.)
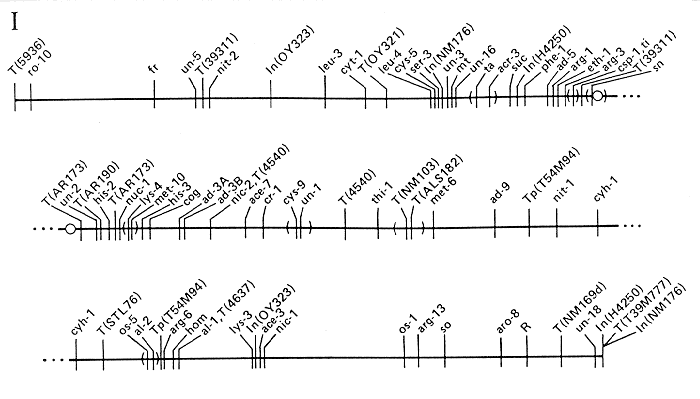
Fig. 1. Linkage group I of N. crassa. The map shows loci whose
order is established with reasonable certainty. Order of markers in parentheses is
uncertain. The centromere is represented as a circle. The order of genes listed below
the map is less well established relative to loci shown on the map; percentages and
fractions indicate recombination among random ascospore progeny, unless stated
otherwise. For detailed information and documentation, see alphabetical entries under
"Information on Individual Loci."
ADDITIONAL LOCI IN LINKAGE GROUP I
- acr-1: Linked to mt (8 to 12%)
- acr-4: Linked to mt, acr-3 (5%)
- acu-4: Right of arg-1 (5/29)
- age-1: Linked to so and aro-8
- age-3: Right of un-18
- amyc: Between ad-5 and the centromere
- aro-7: Between arg-1 (4%) and his-3 (1 to 2%)
- atr-1: Between In(H4250) and T(39311)
- aza-1: Left of mt (23%)
- aza-2: Linked to mt (2%) and aza-1 (39%)
- Bm: Left of nit-2 (30%)
- Ban: Left of mt (14%)
- bs-1: Linked to un-1 (9%), probably to the right
- cnr: Linked to hom (1%), probably to the right
- col-12: Linked to mt (17 to 22%)
- cr-2: Between T(NM103) and al-2 (18%)
- cr-3: Between T(4540) and cr-2
- csh: Between thi-1 (12 to 20%) and ad-9 (5%)
- cy: Linked to ad-5 (1/54), probably to the left
- cya-1: Linked to mt (6%)
- cys-11: Linked to cys-5 (< 1%), probably to the right
- cys-12: Right of ad-9 (12%), linked to al (0/76)
- cys-13: Right of his-3 (2%)
- cyr-4: Between the T(AR173) breakpoints
- cyt-18: Linked to al-2 (10%) and nic-1 (1 to 5%)
- dot: Linked to ad-9 (0/44); right of thi-1 (2%)
- En(pdx): Linked to mi (5%), probably to the left
- erg-4: Linked to al-1 (10%)
- ff-3: Right of os-1 (3%)
- fls: Between nit-1 (5 to 19%) and al-1 (6 to 19%)
- fmf-1: Linked near arg-1
- fpr-5: Left of al-2 (25%)
- fs-3: Left of mt (16%)
- Fsp-2: Right of nic-2 (6%)
- glp-1: Linked to ad-9 (2%), probably to the right
- glp-5: Left of cr-1 (15%)
- gsp: Left of mt (10%)
- gua-1: Probably between his-2 (3%) and cr-1 (3%)
- het-5: Between T(NM103) and the right tip
- ipa: Between mt (20%) and arg-1 (1%)
- lis-1: Between ad-3 (6%) and al-1 (16%)
- lysR: Between his-3 and nic-2
- mb-2: Between cyh-1 (5%) and al-1 (7%)
- mb-3: Linked to cyh-1 (18%), al-1 (2%), and mb-2 (6%)
- mei-3: Between arg-1 (3%) and T(39311)
- mig: Between tre (<< 1%) and ad-9
- mo-1: Linked to mt (9%)
- mo-5: Linked to mt (20%)
- mus-9: Between cyh-1 (18%) and al-2 (6%)
- mus(SC28): Right of al-1 (18%)
- nd: Between the centromere (15%) and al-2 (20%)
- nit-8: Linked to nit-1 (32%) and mt (10%)
- nuh-6: Between the centromere (5%) and nic-2 (4%)
- nuh-8: Right of nic-1
- os-4: Between T(39311) and T(AR173)
- pat: Linked to mt, probably to the right
- prd-3: Near the centromere
- rec-3: Between acr-3 (1 to 2%) and arg-3 (2 to 8%)
- rg-1: Between T(AR173) and lys-4 (1 to 7%)
- rg-2: Linked to mt (15%)
- ro-6: Between the T(4540) breakpoints
- sar-1: Near mt
- sdh-1: Linked to ml (0/13)
- sf: Linked to mt (3%) and cy (3%)
- slo-1: Between mt (14%) and thi-1 (2 to 5%)
- smco-1: Linked to mt (1%) and rg-1 (0/72)
- smco-3: Linked to mt (10%) and al-2 (29%)
- smco-6: Linked to mt (2%)
- sor-4: Linked to phe-1 (< 1%)
- sor(T9): Between mt (6%) and the centromere (5%)
- spco-11: Linked to mt (17%) and mo-5 (18%)
- spco-12: Linked to mt (20 to 35%) and mo-5 (5%)
- ss: Very near nit-2
- ssu-2: Linked to mt (22%) and al-2 (26%)
- ssu-3: Linked to mt (22%) and al-2 (33%)
- st: Between ad-3B (5%) and thi-1 (14%)
- su(bal): Linked to mt (13%)
- su(col-2): Linked to mt (0/837)
- su(met-7)-1: Linked to al-2 (1%)
- su([mi-3])-1: Linked to al-2 (4%)
- su(mtr)-1: Right of his-2 (2%)
- su(trp-3td2)-2a: Linked to al-2 (15%)
- T: Between ad-3A (18%) and al-2
- tet: Linked to acr-3 (2%) and ad-3B (1%)
- tre: Right of met-6 (7%)
- ty-2: Right of al-2
- tyr-2: Linked to cr-1 (2%), probably to the left
- tys: Right of mt (6%)
- uc-2: Linked to mt (0/12 asci)
- un-7: Between T(STL76) and al-1 (3%)
- upr-1: Between mt (2%) and arg-1 (7%)
- ure-4: Linked to his-3 (1%). probably to the right
- uvs-6: Linked to met-6 (< 1%)
- wc-2: Between T(ALS182) and ad-9 (7 to 20%)
- ylo-2: Between ln(H4250) and arg-1 (1%)
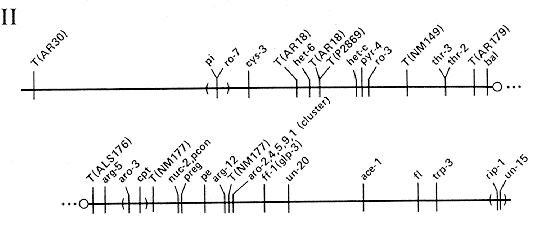
FIG. 2. Linkage group II of N.
crassa. Scale
and conventions as for Fig. 1.
ADDITIONAL LOCI IN LINKAGE GROUP II
- acu-5: Linked to arg-5 (6%) and aro-3 (7%)
- alc-1: Linked to pe (10%)
- aod-2: Linked to arg-5 (7%) and thr-3 (16%)
- col-10: At or near pi
- cot-5: Right of T(P2869); probably left of pyr-4
- cwl: Linked to arg-5 (3%)
- cya-4: Near thr-3
- cyb-3: Left of ro-3 (9%)
- cyl-12: Between thr-3 (38%) and trp-3 (18%)
- da: Linked to thr-3 (3%)
- eas: Linked to rip-1 (1/151), trp-3 (0/71), and fl (1/52)
- en(am)-2: Near pe
- (fs-2) (Tentative linkage cot-5 [14/48])
- Fsp-1: Right of pe (4%)
- glp-2: Between T(ALS176) and T(NM177)
- het-d: Right of fl (25%)
- lp: Right of thr-3 (10%)
- mus-7: Between arg-5 (8 to 12%) and nuc-2 (11%)
- nuh-5: Linked to trp-3 (30%); near T(4637)
- (pcon): (Probably allelic with nuc-2 [0/8541)
- ro-9: Linked to thr-3 (0/63); right of T(NM149)
- scr: Linked to arg-12 (2%), probably to the left
- spco-14: Linked to arg-5 (7%)
- su([mi-1])-3 Linked to fl (31%)
- su([mi-1])-4: Linked to fl (22%) and arg-5 (40%)
- su([mi-1])-10: Linked to arg-5 (29%) and fl (24%)
- tng: Between T(NM149) and arg-5 (6 to 14%)
- tu: Between pe (8%) and fl (19%)
- uc-1: Linked to pyr-4 (31%)
- ure-3: Between arg-12 (7 to 12%) and ace-1 (14%)
- xdh-1: Linked to pe (14%)
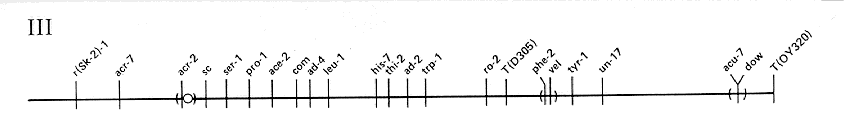
FIG. 3. Linkage group III of N. crassa. Scale and conventions as
for Fig. 1.
ADDITIONAL LOCI IN LINKAGE GROUP III
- acon-2: Linked to vel (6%) and tyr-1 (9 to 14%)
- acr-6: Linked to shg (0/368)
- aza-3: Linked to trp-1 (14%)
- col-16: Linked to leu-1 (1%) and pro-1 (10%)
- cum: Linked to acr-7 (8 to 18%), acr-2 (18%), and Sk-2
- cya-7: Linked to ad-4 (25%)
- cyt-8 : Linked near ad-4
- erg-3: Linked to dow (10%), probably to the left
- ff-5: Between pro-1 (2%) and met-8 (1%)
- ff-6: Linked near ty-1
- fpr-3 : Linked to trp-1 (1%) and thi-2 (5%)
- gluc-1: Linked to dow, (10%)
- het-7: Between T(D305) and the right tip
- mei-4: Linked to leu-1 (12%), probably to the left
- met-8: Between ff-5 (1 to 4%) and ad-4 (4%)
- mo-4: Right of leu-1 (8%)
- nit-7: Between trp-1 (26 to 32%) and dow (45%)
- nuh-1: Between leu-1 (4%) and nuh-2 (< 1 %)
- nuh-2: Between nuh-1 (< 1 %) and trp-1 (11%)
- ota: Linked to pro-4 (4%); between ad-4 (15%) and tyr-1 (14%)
- pgov: Linked to tyr-1 (1 to 4%), probably to the right
- prd-1: Linked to acr-2 (5%) and pro-1 (20%)
- pro-4: Linked to thi-2 (0/78) and ota (4%)
- ser-5: Linked to trp-1 (1%) and ser-1 (12%)
- shg: Linked to trp-1 (7%)
- Sk-2: Left of his-7 (25%)
- Sk-3: Left of his-7 (11%)
- sor-3 : Linked to ad-4 (7%)
- spg: Linked to sc (< 1 %) and thi-4 (0/103)
- su([mi-1])-1: Linked to ad-4 (14%)
- su(trp-3td2)-2: Linked to leu-1 (22%)
- thi-4: Linked to acr-2, spg, and sc (< 1 %)
- trk: Linked to leu-1 (0/92)
- ty-1: Linked to tyr-1 (0 to 6%) and dow (21%)
- un-6: Right of sc (21%) and acr-2 (6 to 20%)
- un-14: Right of acr-2 and thi-4 (8 to 20%); left of leu-1 (5%)
- un-21: Between acr-2 and un-6
- uvs-4: Left of ad-4 (4%)
- uvs-5: Linked to vel (1%)
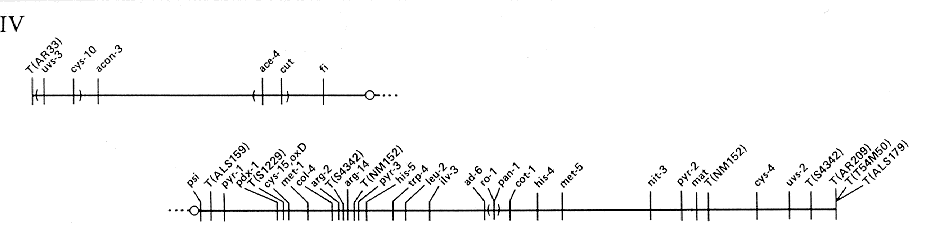
FIG. 4. Linkage group IV of N. crassa.
Scale and
conventions as for Fig. 1.
ADDITIONAL LOCI IN LINKAGE GROUP IV
- acu-2: Between leu-2 (11%) and pan-1 (6%)
- ads: Linked to col-4
- aod-1: Left of trp-4 (23%)
- bd: Right of pan-1 (2%)
- car: Linked to cys-10 (1%)
- cel: Linked to pan-1 (1%)
- chol-1: Linked to ad-6 (1%), probably to the right
- coil: Between arg-2 (2%) and leu-2 (12%)
- col-1: Linked to pan-1 (0/47 asci) and cot-1 (3%)
- col-5: Linked to cot-1 (1%), probably to the right
- col-6: Linked to the centromere (0/28 asci) and pan-1 (21%)
- col-8: Linked to pan-1 (4 to 13%)
- cot-3: Left of pan-1 (16 to 25%); probably right of arg-2
- crib-1: Linked to met-1 (6%)
- cya-5: Right of pan-1 (2%)
- cya-6: Right of pan-1 (2%)
- cys-14: Linked to cot-1 (21%)
- cyt-5: Left of trp-4 (9%)
- cyt-19: Linked to cyt-5 (1/201)
- an : Linked to mat (1%); right of pyr-2 (4%)
- fdu-2 : Right of cys-4 (2%)
- fld : Left of his-5 (2%)
- gpi-1: Linked to ad-6 (10%)
- gua-2: Linked to cot-1 (5%)
- gul-3: Linked to cot-1 (10%) and pyr-2 (7%)
- int: Linked to pan-1 (0/50)
- le-1: Linked to pan-1 (1 to 2%); right of cot-1 (14%)
- med: Linked to met-5 (5%) and pan-1 (8%)
- mei-1: Linked to arg-2 (< 1 %), probably to the right
- met-2: Linked to ilv-3 (0/129); between trp-4 (6%) and pan-1 (4%)
- mod(sc): Linked to pan-1 (17%)
- moe-3: Left of pan-1 (17 to 25%) and bd (18%)
- mtr: Between pdr-1 (2%) and col-4 (1%)
- mus-8: Linked to mtr (1%) and pdx-1 (6%)
- nada: Left of ad-6 (18%)
- nit-4: Linked to col-4 (2%), probably to the right; left of pan-1 (6 to 27%)
- nit-9: Right of nit-4 (9%)
- os-2: Right of cot-1 (4%)
- pf: Linked to mat (3%); right of pyr-2 (2%)
- pho-3: Linked to pan-1 (< 1 %)
- pmb: Right of uvs-2 (8%)
- Pt: Right of T(S1229) and pdx-1 (2%); left of col-4 (2%)
- rib-2: Between T(S4342) and chol-1
- rol-1: Linked to pdx-1 (0/88)
- ser-4: Right of arg-2 (< 1 %)
- smco-4: Linked to pan-1 (8%)
- smco-8: Linked to pan-1 (1 to 7%) and smco-4 (30%)
- smco-9: Linked to pan-1 (5%), smco-4 (2%), and smco-8 (13%)
- spco-8: Linked to pan-1 (23%)
- su([mi-1])-14: Linked to arg-2 (14%)
- thi-5: Linked to pan-1 (1%)
- tol: Linked to trp-4 (1%), probably to the left
- uc-5: Right of cot-1 (32%)
- un-8: Linked to pyr-1 (0/47); between T(ALS159) and col-4 (5%)
- un-12: Linked to col-4 (0/73) and pdx-1 (5%)
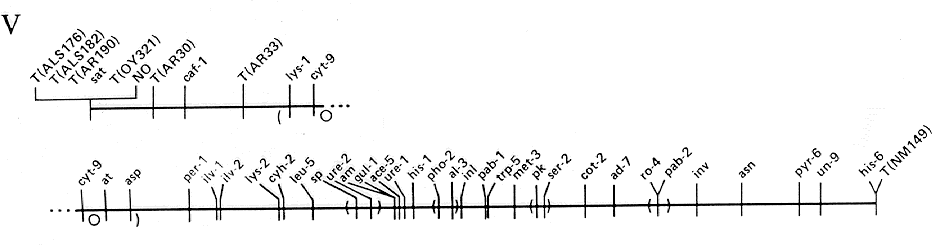
FIG. 5. Linkage group V of N. crassa. Scale and conventions as for Fig.
1.
ADDITIONAL LOCI IN LINKAGE GROUP V
- acu-1: Right of asn (21%)
- acu-3: Between ini (7%) and asn (20%)
- arg-4: Between sp (1 to 11%) and ini (2 to 4%)
- cl: Right of pk (< 1 to 2%)
- col-9: Between int (16%) and asn (5%)
- cot-4: Between rol-3 (5%) and ini (10%)
- cya-2: Linked to al-3 (3%) (indicated to the right)
- cyb-1: Between al-3 (24%) and his-6 (10%)
- dgr: Linked to caf-1 (8%)
- en(am)-1: Between am-1 (8%) and inl (1%)
- erg-1: Between pk (2%) and asn (9%)
- erg-2: Left of inl (6%)
- fpr-1: Linked to cyh-2 (< 1%)
- fpr-4: Right of inl (11%)
- gln-1: Linked to inl (2%), probably to the right
- glp-6: Left of inl (30%)
- gran: Linked to pl (0/75); between pab-2 (1 to 8%) and his-6 (8 to 27%)
- hgu-4: Between cyh-2 (7%) and ure-2 (10%)
- lasc: Right arm
- lis-3: Right of inl (4%)
- md: Between sh (3%) and sp (9%)
- Mei-2: Linked near inl
- mus-11: Linked to his-6 and pab-2
- mus(SC17): Left of inl (27%)
- nap: Linked to int (15%); right of ure-2 (32%)
- ndc-1: Left of arg-4 (2%)
- nmr-1: Between am (3 to 7%) and gln-1 (4 to 10%)
- nuh-3: Between cyh-2 (4%) and al-3 (17%)
- nuh(23): Linked to nuh-3 (6%)
- pl: Linked to gran (0/75)-. between asn (1 to 9%) and his-6 (16%)
- prd-2: Right arm
- pro-3: Linked to inl and pab-1 (0/74); between his-1 (4%) and pk (2 to 6%)
- rec-1: Between ro-4 (7%) and asn (5%)
- rec-2: Between sp and am
- rol-3: Between ilv-1 (2%) and cot-4 (5%)
- scon: Probably linked to VR; right of his-6
- scot: Between al-3 (7%) and his-6 (11%)
- sh: Between ilv (4 to 7%) and md (3%)
- smco-6: Linked to asn (6%); near pyr-6
- smca-7: Linked to rol-3 (0/154); right of ilv-1 (2%)
- spco-9: Linked to asn (6%); right of met-3 (18%)
- spe-1: Linked to cyh-2 (2%); left of inl (14%)
- su(ile-1): Right of met-3 (4%)
- su([mi- 1])-f: Left of inl (10%)
- ts: Linked to inl (4%)
- uc-4: Between inl (12%) and his-6 (11%)
- udk: Left of uc-4 (29%)
- un-11: Linked to al-3 (0/48)
- un-19: Linked to al-3 (9%) and un-11 (14%)
- val: Linked to ilv-2 (0/135)
- wa: Linked to inl (6%)
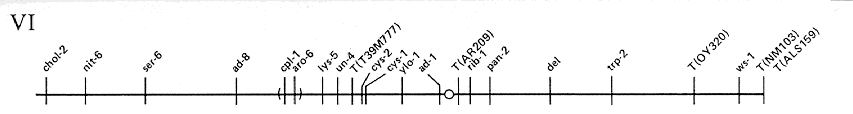
FIG. 6. Linkage group VI of N. crassa.
Scale and
conventions as for Fig. 1.
ADDITIONAL LOCI IN LINKAGE GROUP VI
- acu-6: Left of cys-1 (3%)
- age-2: Right of ws-1
- Bml: Linked to cys-2 (2%) and ylo-1 (2 to 3%), probably between
- chr: Between chol-2 (10%) and pan-2
- cpc-1: Right of ylo-1 (1-3%)
- cya-3: Between chol-2 (10%) and cyt-2 (10%)
- cys-2: Between un-4 (4%) and cys-1 (1 to 3%)
- cyt-2: Between cya-3 (10%) and lys-5 (6%)
- edr-1: Linked to ad-1 and pan-2 (0/125)
- edr-2: Left of ad-1 (19%)
- fpr-6: Between pan-2 and trp-2
- glp-4: Between ad-1 (0 to 2%) and rib-1 (3 to 4%)
- gul-5: Linked to trp-2 (10%)
- het-8: Between chol-2 (19%) and ad-8 (12%)
- het-9: Between T(AR209) and the right tip
- lis-2: Between chol-2 (11%) and trp-2 (25%)
- mod-5: Linked to trp-2, near the centromere (3%)
- moe-2: Linked to trp-2 (14%), probably to the left
- mts: Right of ylo-1 (< 1 %)
- sor-1: Left of ylo-1 (3%)
- spco-7: Linked to ad-1 (0/65), between ylo-1 (4%) and T(AR209)
- spco-13: Linked to trp-2 (16%) and the centromere (1/10 asci)
- ssu-7: Between ad-8 (8%) and ylo-1 (14%)
- su(mtr26): Linked to trp-2, in the right arm
- su(trp-5): Near aro-6 (allelic?)
- un-13: Linked to ylo-1 (2%) and lys-5 (4%)
- un-23: Right of trp-2 (5 to 27%); left of ws-1
- ws-2: Linked to trp-2 (2%) and ylo-1 (16%)

FIG. 7. Linkage group VII of N. crassa. Scale and conventions as
for Fig. 1.
ADDITIONAL LOCI IN LINKAGE GROUP VII
- adh: Linked to do (0/53), spco-4 (4%), and nic-3 (11%)
- aga: Between wc-1 (2%) and arg-10 (24 to 27%)
- aln-1: No data
- ars: Right of thi-3 (2 to 5%); left of ile-1 (1%) and met-7 (< 1%)
- bn: Linked to thi-3 (2%); right of T(T54M50)
- col-2: Linked to met-7 (I%)
- col-3: Linked to met-7 (0/93) and wc-1 (1%)
- col-17: Linked to nt (14%) and spco-5 (6%)
- cyt-6: Linked to wc-1 (2%) (indicated left)
- cyt-7: Linked to nic-3 (18%) (indicated left)
- gul-4: Linked to nic-3 (17%)
- het-10: Between T(5936) and the right tip
- hlp-1: Between sfo (1 to 9%) and nt (28 to 37%)
- hlp-2: Between hlp-1 (8 to 25%) and nt (29%)
- ile-1: Between ars (1%) and wc-1 (< 1 to 2%)
- kyn-1: Linked to nic-3 (30%) and wc-1 (200/v)
- le-2: Linked to met-7 (7%)
- mb-1: Linked to nic-3 and wc-1 (23%)
- met-1: Left of thi-3 (27%)
- mo-2: Linked to nt (29%)
- mus-10: Right of met-7 (7%)
- oli: Linked to frq (< 2%)
- rol-2: Linked to met-7 (0/298)
- sfo: Between thi-3 (6%) and htp-1 (1 to 9%)
- slo-2: Left of met-7 (2%)
- sor-2: Linked to nt (31%)
- spco-5: Linked to col-17 (6%) and nt (20%)
- spco-6: Linked to do (10%), spco-5 (8%), and nt(20%)
- ssu-1: Between met-7 (14%) and su(trp-3td201)-1 (10%)
- ssu-4: Between nic-3 (28%) and met-7 (20%)
- su([mi-1])-5: Left of nic-3 (23%)
- sti(trp-3td201)-1: Right of ssu-1 (10 to 13%); left of arg-10(7%)
- su(trp-3td201)-2: Linked to su(trp-3td201)-1, probably to the left
- ud-1: Between met-7 (27%) and arg-10 (10%)
- un-22: Linked to met-7 (1%)
- van: Left of nic-3 (4%)






