With the development of molecular biology techniques and their application to the analysis of cellular events, the isolation of total nucleic acids in Aspergillus nidulans for Southern and Northern hybridization has become routine. The current methods of nucleic acids isolation in A. nidulans, however, are relatively tedious and limited to a small number of samples each time. For example, the method for nucleic acid isolation of Oakley et al. (1987 Gene 61:385-399) requires the grinding of mycelia, either in frozen or lyophilized form, before extraction with phenol. This method requires a relatively large amount of mycelium and up to a day of preparing the mycelium before the actual extraction of nucleic acids. To increase the efficiency so that many samples can be studied at the same time, we have modified a method for nucleic acids isolation from Saccharomyces cerevisiae (Elder et al. 1983 Proc. Natl. Acad. Sci. (USA) 80:1194-1198) and adapted it to A. nidulans.
Experimental Procedures
The mycelia were harvested by filtration with a Buchner funnel and their wet weights were determined. One-tenth of a gram of mycelium was weighed out and resuspended in an Eppendorf tube with 0.2 ml buffer containing 0.2 M Tris-HCl, (pH 7.5), 0.5 M NaCl, 0.01 M EDTA, and 1% SDS. 0.4 gm of acid washed glass beads (0.3-0.4 micron) and 0.2 ml of phenol:chloroform (1:1 v/v) were added to the mycelial suspension. The mixture was then vortexed at top speed for 6 min. An additional 0.3 ml of buffer solution and 0.3 ml phenol:chloroform were added to the vortexed mixture. The solution was mixed by vortexing for a few seconds and then centrifuged in a microfuge for 30 sec to separate the phases. The aqueous phase was removed and re-extracted in a new tube with 0.3 ml more of phenol:chloroform. This mixture was centrifuged for another 30 sec and the aqueous phase was removed and placed into a new tube. The tube was then filled with 95% ethanol and stored at -20°C for 30 min or longer. The nucleic acids were pelleted in the microfuge tube by centrifugation for 5 min, washed with 300 ul of 70% ethanol, and then dried in a Speed Vac. The nucleic acid pellet was dissolved in 100 ul of sterile distilled water.
Results and Discussion
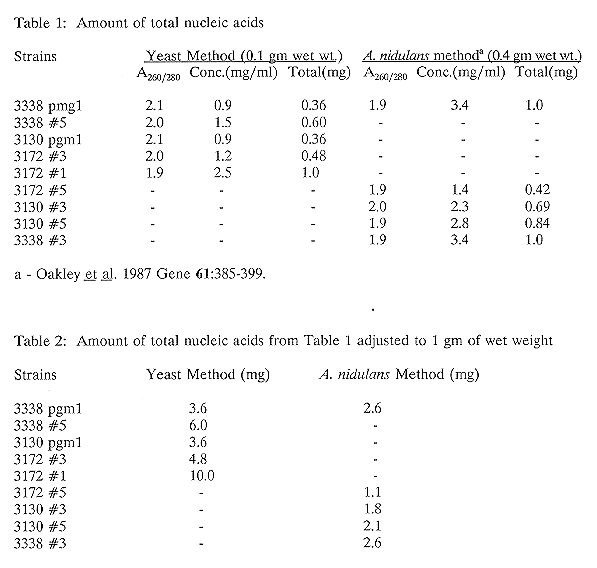
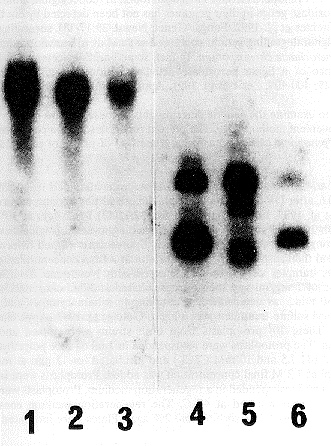
We have isolated total nucleic acids from 10 different preparations of mycelium from A. nidulans transformants. The transformants were obtained by transformation with a pAL5 plasmid as modified and provided by S. Osmani (Waring, May, and Morris, 1989 Gene 79: 119) containing a Neurospora crassa endo-exonuclease cDNA (T. Chow, unpublished result). Five of the preparations were isolated with the method described by Oakley et al. (1987) and the other five were isolated with the new method described above. One of the preparations in each method is of the same transformant, to eliminate the possibility of strain differences. As Table 1 indicates, the A260/A280 ratio for all the preparations are the same, suggesting the major components are nucleic acids. When the amount of the total nucleic acids isolated is expressed as milligrams of nucleic acids recovered per 1 gm of wet weight, the data indicate that the yeast method gives a much higher yield than the method described by Oakley et al. (1987) (Table 2). To confirm that the quality of the nucleic acids isolated with our method is as good as that obtained with the Oakley et al. method, we subjected the nucleic acids obtained by both methods to agarose gel electrophoresis after treatment with the restriction enzyme EcoRI, then transferred to Amersham Hybond-N membrane and hybridized with the radioactive labelled N. crassa endo-exonuclease cDNA. The results of the Southern hybridization (Figure 1) shows similar hybridization intensity for the nucleic acids prepared with both methods, suggesting the quality of the isolated nucleic acids from the two preparations is the same. (Supported by NCIC, CRS and FRSQ)


Figure 1. Southern hybridization of Aspergillus nidulans total nucleic acids isolated from transformants of a
pAL5 plasmid containing a Neurospora crassa endo-exonuclease cDNA with the 32P-labeled N. crassa endo-exonuclease cDNA.
Lane 1, nucleic acids from transformant 3130 #3 isolated with the method described by
Oakley et al. 1987 (1 µg)
lane 2, nucleic acids from transformant 3172 #3 isolated with the
method described above (1 µg)
lane 3, nucleic acids from transformant 3172 #1 isolated with the method
described above (0.5 µg)
lane 4-6, same sample as in lane 1-3, but digested with the restriction enzyme EcoRI.