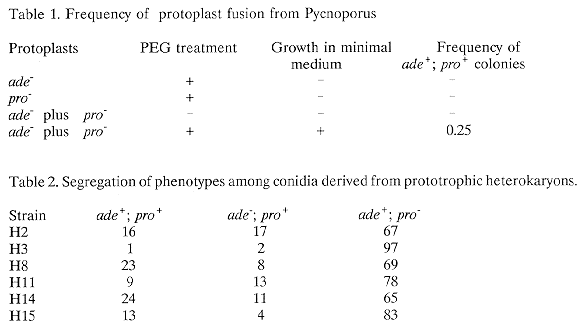
Strains H2, H3, H8, H11, H14 and H15 correspond to heterokaryons obtained by protoplast fusion. Values are percentages of phenotypes indicated in the table.
White-rot basidiomycetes are known as very powerful degraders of wood components. The relative enzyme activities during the degrading process may vary between different species. Several Pycnoporus strains have been found to reduce lignin, although the presence of lignin peroxidase genes in their genomes has not been detected by dot blot hybridization analysis (Cifuentes et al. 1992. Fungal Genet. Newsl. 39:17-18), suggesting the existence of a different lignin degrading system compared with what is known in other basidiomycetes such as Phanerochaete chrysosporium. In fact, some basidiomycetes seem to degrade lignin in the absence of a lignin peroxidase activity (Waldner et al. 1988. Appl. Microbiol. Biotechnol. 29: 400-407; Perie et al. 1991. Appl. Environ. Microbiol. 57: 2240-2245).
In order to examine the genetic determinants of these systems it is necessary to explore the most convenient methodology. The present report describes a complementation genetic analysis in Pycnoporus cinnabarinus using a method of fusion of protoplasts derived from conidia.
P. cinnabarinus mutant strains ade- and pro- were obtained from a haploid wild type strain, Pc470.6, after UV irradiation. Mycelia of both strains were grown in solid PC medium (Cifuentes et al. 1989. Fungal Genet. Newsl. 37:12-13) for 7 days at 30°C. Conidia were collected with 10 ml of sterile water. The remaining mycelial fragments were removed by filtration through three layers of cheesecloth. Spores were washed twice with 10 ml of 1.2 M sorbitol and then suspended in 1.2 M sorbitol at a final concentration of 10(8) spores/ml. One milliliter samples of spores were treated with Novozyme 234 (Sigma) at a final concentration of 2 mg/ml and then were incubated at 37°C until 100% protoplasts were obtained (150 min.), as determined by counting in a hemocytometer after 0.3 and 0.5% sodium dodecyl sulfate treatment according to Gold et al. 1983 (Appl. Environ. Microbiol. 46:260-263). Then, 10(7) protoplasts from both strains were mixed and sedimented by centrifugation. The protoplasts were resuspended in 1 ml of 40% polyethylene glycol (in 10 mM Tris-HCl pH 7.5 and 10 mM CaCl2) and incubated for 15 min at room temperature. Then, sorbitol at 1.2 M final concentration was added. Protoplasts were washed twice with 1.2 M sorbitol and resuspended in a regeneration medium. Protoplasts were plated in 20 ml of regeneration agar warmed at 45°C. The regeneration medium consisted of Vogel's minimal medium plus 1.2 M sorbitol and 2% agar. Plates were incubated at 30°C for 5 to 8 days.
Table 1 shows the results of protoplast fusion experiments as measured by complementation on plates of minimal medium. The frequency of fusion was determined as the ratio between the number of colonies formed on minimal medium and colonies formed on the same medium supplemented with adenine and proline. When polyethylene glycol (PEG) was omitted from the mixture, complementation did not occur.
Table 2 shows the results of a segregation analysis of conidia from six heterokaryons obtained from minimal medium after protoplast fusion. Conidia obtained in these heterokaryons were suspended in sterile water at a final concentration of 5 x 10(3). 0.1 ml samples were then plated in minimal medium alone and minimal medium containing adenine or proline as a supplement. All heterokaryons showed conidia segregation of both ade- and pro- markers from parental strains.

Strains H2, H3, H8, H11, H14 and H15 correspond to heterokaryons obtained by protoplast
fusion. Values are percentages of phenotypes indicated in the table.
Because it is difficult to perform classical genetic studies in P. cinnabarinus, its genome is largely undescribed. An alternative way could be the application of parasexual genetic analysis. Our results indicate that protoplast fusion is an efficient method to conduct genetic analysis in P. cinnabarinus and may be used to study lignin degrading variations in different mutants strains affecting phenol oxidase activities obtained in our laboratory. On the other hand, it will be possible to take advantage of protoplasts of auxotrophic strains (ade-) for genetic transformation experiments with DNA of wild-type strains.