Complementation of Cochliobolus heterostrophus trp- mutants produced by gene
replacement
P.G. Mullin, B.G. Turgeon and O.C. Yoder - Department of Plant Pathology,
Cornell University, Ithaca NY 14853
Transformation systems for most filamentous fungi are based on selection
for drug resistance. This strategy is advantageous becasue wild-type strains,
including isolates collected directly from the field, can be used as
recipients in transformation experiements. Drug resistance as a selection
strategy is limited for those fungi which are insensitive to most drugs since
the number of selectable markers available for sequential indroduction of
genes is reduced. An alternative approach is complementation of an auxotroph
with a cloned gene encoding the missing enzyme, followed by selection for
prototrophic growth. Auxotroph complementation is widely used with
genetically developed fungi, but most economically important species lack the
requisite auxotrophic strains. In this study, we produced an auxotroph by
mutating the native copy fo the tryptophan synthase, TRP1 (Turgeon et al. 1986
Gene 42:79-88), of the plant-pathogenic fungus Cochliobolus heterostrophus.
The resulting trp- strain was readily transformed to prototrophy using TRP1.
This adds auxotroph complementation to the drug resistance [hygromycin B and
the hygB gene (Turgeon et al. 1987 Mol. Cell. Biol. 7:3297-3305), bialaphos
and the bar gene (Straubinger et al. 1992 Fungal Genet. Newsl. 39:82-83)] and
substrate utilization [acetamide and the amdS gene (Turgeon et al. 1985 Mol.
Gen. Genet. 201:450-453)] systems available for transformation of C.
heterostrophus.
The sequence of C. heterostrophus TRP1 has been deposited (EMBL Data
Library Accession Number X70035). There is an open reading frame (Figure 1)
extending from bp 544 to bp 2989 (including the stop codon), interrupted by an
apparent intron (140 bp) from position 863 to 1002. At the amino acid level,
the gene has 62% and 64% identity to the corresponding genes from Aspergillus
nidulans (trpC) and Neurospora crassa (trp-1).
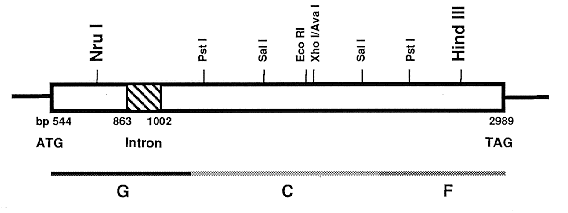
Figure 1. Structure of the C. heterostrophus TRP1 gene, which encodes a trifunctional polypeptide that
performs three separate enzymatic conversions in the tryptophan biosynthetic pathway (G=step 1, glutamine
amidotransferase; C=step 4, indole glycerol phosphate synthase; F=step 3, phosphoribosyl anthranilate
isomerase); the direction of transcription is from left to right. The TRP1 sequence is similar to that of trpC
from Aspergillus nidulans and trp-1 from Neurospora crassa, except that only the C. heterostrophus gene
contains an intron. Six-base-pair restriction enzyme recognition sites are shown; large bold letters indicate
sites of introduced frameshift mutations. Base pair numbering refers to the sequence as listed in the EMBL
database (Acc. no. X70035)
Sequence and restriction enzyme site data were used to construct pTK2 (Figure
2), a vector designed for introduction of a point mutation into TRP1. It is based on
plasmid pGB2 (Churchward et al. 1984 Gene 31:165), which has no homology to pBR322
and can be used when interaction with pBR322-based vectors is to be avoided. pGB2
was digested with SmaI and a 3.8 kb BamHI-PvuII fragment containing TRP1 from
pChTrp24B (Turgeon et al.1986 Gene 42:79-88) was inserted, eliminating the SmaI site.
The new plasmid, pETE3, was digested with NruI, which cuts once in the coding region
(bp 734, in the G domain) of TRP1; SmaI linkers were added and the plasmid religated.
Insertion of linkers into the NruI site introduced a frame-shift mutation, which was
confirmed by restriction mapping and sequence analysis. This plasmid, pTKG, was then
partially digested with HindIII and end-filled. Restriction anayisis identified a plasmid,
pTKGF, which contained a mutation in the HindIII site at the 3' end of the coding
region (bp 2653) of TRP1 (Fig. 2). pTKGF was digested with HindIII, blunt-ended and
ligated to NruI digested pDH25 (Cullen et al.1987 Gene 57:21-26). The final
construction, pTK2, contained the hygromycin B phosphotransferase gene from
Escherichia coli with 5' and 3' regulatory sequences from A. nidulans trpC and the C.
heterostrophus trp1 gene with frame shift mutations in the 5' and 3' regions of the ORF.
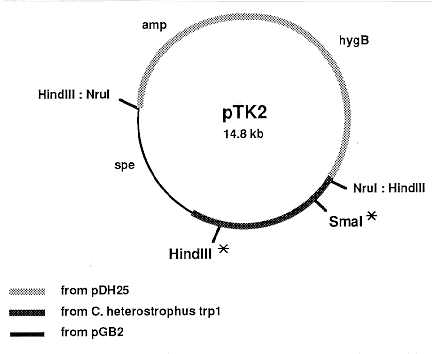
Figure 2. Structure of pTK2, constructed as described in the text. Asterisks indicate positions of point
mutations introduced into the TRP1 ORF. amp and spe: E. coli genes for resistance to ampicillin and
spectinomycin; hygB: E. coli gene for resistance to hygromycin B, fused to A. nidulans trpC initiation and
termination signals for selection in fungi.
A trp1 mutant strain of C. heterostrophus was constructed by two-step gene
replacement. Protoplasts of wild-type strain C3 (tox1-, MAT1-2; ATCC 48330) were
transformed (Turgeon et al. 1987 Mol. Cell. Biol. 7:3297-3305) with pTK2. Seventy two
hygromycin B resistant transformants were purified by single conidiation and screened
for ability to grow on complete medium in the absence of supplemental tryptophan.
Nearly 20% were unable to grow on unsupplemented complete medium, but grew
normally (as compared with the untransformed strain C3) on medium containing either
tryptophan (4 mM final concentration) or indole (25 ug/ml). These Trp- transformants
retained their resistance to hygromycin B. Southern analyses of genomic DNAs from the
transformants revealed both homologous and ectopic integrations. Transformants with
signals which matched those predicted for homologous integration of a single copy of
pTK2 at TRP1 (which resulted in a recombinant chromosome with two copies of trp1:
one carrying the 5' mutation and one carrying the 3' mutation, separated by vector
sequences) were crossed to C. heterostrophus strain C2 (alb1 Tox1+ MAT1-1; ATCC
48329); the goal was to evict one copy of trp1 and the invervening vector DNA. Mating
medium was supplemented with indole (25 ug/ml); no viable ascospores were found
when mating medium was not supplemented or contained 4 mM tryptophan. One of the
trp1- hygBR transformants, pTK2.4.4.4, yielded progeny that were predominantly parental
but also included progeny which were trp1-, hygBS. Southern analysis of one of these
ascospore isolates, D1.51 (alb1, hygBS, MAT1-1, Tox1+, trp1-) confirmed that one copy of
trp1 and all vector sequences had been evicted. The chromosome of this isolated was
identical to that of wild type except that the 3' HindIII site in the F domain of TRP1 was
missing, as expected.
The trp1- mutant of C. heterostrophus was transformed to prototrophy with TRP1.
Protoplasts from TMHS24 (a pigmented backcross progeny derived from D1.51) were
treated with plasmid DNA carrying TRP1 (e.g. pChTRP24B). Southern analyses of
genomic DNAs isolated from transformants showed both homologous and ectopic
integrations of transforming plasmid resulting in prototrophy.
Availability of two-step gene replacement for C. heterostrophus means that it
should be possible to assess the biological function of any cloned single-copy gene from
this fungus by replacing the wild-type allele with a copy that has been specifically altered
in vitro. In addition, complementation of the trp1- auxotroph with the cloned TRP1 gene
provides an alternative transformation strategy for C. heterostrophus. Approximately 70%
of all integration events are the result of homologous integration of the transforming
DNA at TRP1. For comparison, homologous integration with promoter 1 from C.
heterostrophus (Turgeon et al.1987 Mol. Cell. Biol. 7:3297-3305) is approximately 85%
when 1 kb of homologous DNA is carried on the transforming plasmid. Random
integration occurs when the transforming plasmid contains no C. heterostrophus DNA
(Turgeon et al. 1985 Mol. Gen. Genet. 201:450-453).

