Strains for identifying and studying individual vegetative (heterokaryon) incompatibility loci in Neurospora crassa.
D. D. Perkins1, J. F. Leslie2 and D. J. Jacobson3 - 1Department of Biological Sciences, Stanford University, Stanford, California 94305-5020; 2Department of Plant Pathology, Kansas State University, Manhattan, Kansas 66506-5502; and 3Department of Botany and Plant Pathology, Michigan State University, East Lansing, Michigan 48824-1312
Genetic and molecular studies of vegetative incompatibility are proceeding in several Neurospora labs. The purpose of this note is to present an expanded list of strains in the Fungal Genetics Stock Center that are potentially useful when partial diploids are employed to identify different alleles at any of the 11 known het loci of N. crassa. Some of the strains are newly deposited in FGSC. Others have previously been listed under other categories in the stock list.
Wild populations of N. crassa are polymorphic for het genes (Mylyk 1976 Genetics 83:275-284). Laboratory strains, which have come from varied lineages, frequently differ from one another in het genotype. This polymorphism and the multiplicity of het loci often make it difficult to use heterokaryon tests for genetic analysis, because failure to complement may result from allelic differences at any one of numerous het loci. Extraneous het genes other than at the locus of interest will usually not be a problem when duplications (partial diploids) are used.
Duplications of known content can be obtained for defined chromosomal segments in progeny of crosses heterozygous for insertional or terminal translocations (see Perkins and Barry 1977 Advan. Genet. 19:133-295). Because the duplications exist in an otherwise haploid genome, they make it possible to identify individual vegetative incompatibility (het) genes and to study them one by one without the necessity of making strains isogenic or homozygous for other het genes located outside the duplicated segment. If the translocation and normal-sequence parents differ with respect to alleles at a het locus within the duplication, then duplication progeny heterozygous for the included incompatible allelic combination display a characteristic inhibited growth with abnormal morphology and pigment (Newmeyer and Taylor 1967 Genetics 56:771-791; Perkins 1975 Genetics 80:87-105; Mylyk 1975 Genetics 80:107-124, 83:275-284). These heterozygous (hetx/hety) duplications are clearly distinguishable from homozygous (hetx/hetx) or (hety/hety) duplication strains, which are usually phenotypically normal or nearly so.
Heterokaryon incompatibility has been shown to correspond with phenotypic abnormality of heterozygous duplications for het genes at seven loci - (mating type [Newmeyer 1970 Can. J. Genet. Cytol. 12:914-926], het-c, d, -e, -5, and -8 [see Mylyk 1976 Genetics 83:275-284], and het-6 [D. J. Jacobson unpublished]). Three loci, het-7, -9, and -10, have been defined solely on the basis of their behavior in duplications. Presumably unlike alleles at these three loci are also heterokaryon incompatible, although this has not been tested because strains are not available that are known to differ only at the het locus in question but not at other loci. het-i has been defined only by behavior in heterokaryons; it differs from other het genes in such a way that incompatibility of different het-i alleles may not be detectable in duplications (Pittenger and Brawner 1961 Genetics 46:1645-1663). Stocks with forcing markers are available for heterokaryon tests of het-c, -d, and -e in eight genotype combinations (prepared by L. Garnjobst and J. Wilson). These are listed in part VII.D.1 of the FGSC Stock List. Strains in this set are probably identical to the Oak Ridge (OR) wild type and its derivatives at het loci other than het-c, -d, and -e. OR strains are het-C het-d het-e het-i het-5OR het-6OR het-7OR het-8OR het-9OR het-10OR. A few wild strains carry tol, a recessive suppressor of the het incompatibility associated with mating type, but OR and most other N. crassa strains are tol+.
Genetic evidence suggests the existence of multiple alleles at two loci-het-c and het-8 (Howlett, Leslie, and Perkins, 1993 Fungal Genet. Newsl. 40). However, multiple allelism could be simulated by two alleles at each of two closely linked het loci, and this alternative has not been ruled out.
The listing that follows (Table 1) is comprised of reference strains and strains with relevant linked markers, both in normal sequence and in the sequence of rearrangements capable of generating duplications that include the locus in question. Different het alleles are denoted by superscripts based on the wild strains of origin or on a laboratory reference strain, for example AD - Adiopodoumé, CR - Costa Rica, HO - Houma, LI - Liberia, OR - Oak Ridge, PA - Panama. Symbols for het-c, -d, -e, and -i are exceptions, with unraised capital or small letters used to specify the first two alleles, e.g. het-D, het-d. These, together with mating type, were the first het loci to be identified. Map relations of the markers and loci are shown in Figure 1. Updated versions of the list will appear in the FGSC Stock List (Part VII.D, Special-Purpose Stocks). (Contribution No. 93-355-A from the Kansas Agricultural Experimental Station, Manhattan.)
Table 1. Strains for studying individual het-loci of N. crassa
FGSC No.
Genotype A a
het-c (IIL) (all are het-6OR)
het-C (OR wild types) 2489 4200
het-c 7335 7336
het-C pyr-4 4030 4031
het-c pyr-4 7145 7146
cot-5 het-C 3560 3561
cot-5 het-c 7447
cot-5 het-C pyr-4 thr-2 7355 7356
T(IIL VR)NM149 het-C 3879 3880
T(IIL VR)NM149 het-c 1483 1482
T(IIL VR)NM149 het-C pyr-4 3136
T(IIL VR)NM149 het-C ro-3 2011 2012
het-cAD 430 2614
het-cAD pyr-4 thr-2 7313
T(IIL VR)NM149 het-cAD 2191 2192
T(IIL VR)NM149 het-cAD pyr-4 7314 7315
het-d (IIR) (all are het-C)
het-D (RL wild types) 2218 2219
het-d (OR wild types) 2489 4200
T(IIR VL)ALS176 het-D 2414 3014
T(IIR VL)ALS176 het-d 3013 2415
T(IIR IVR)OY337 het-D 7472 7473
T(IIR IVR)OY337 het-d 3666 3667
FGSC No.
Genotype A a
het-e (VIIL)
het-E (RL wild types) 2218 2219
het-e (OR wild types) 2489 4200
T(VIIL IVR)T54M50 het-E 2603 2604
T(VIIL IVR)T54M50 het-e 2466 2467
T(VIIL IVR)T54M50 het-e nic-3 3132 3133
het-i (I or II by linkage to translocation 4637 al-1)
het-I al-2 nic-1 7343
het-i al-2 nic-1 7344
het-I T(I;II)4637 al-1; pan-1 7342
het-i (ST74A, 8-1a) 262 988
het-5 (IR)
het-5PA (Panama CZ30.6) 1131
arg-13 het-5PA (b11 OR) 7345
thi-1 ad-9 nit-1 het-5PA (b10 OR) 7348 7349
T(IR VIR)NM103 het-5PA (b4 OR) 7346 7347
het-5OR (OR wild types) 2489 4200
T(IR II)MD2 het-5OR 3826 3827
T(IR VIR)NM103 cyh-1 al-1 arg-13 R het-5OR 3135
het-6 (IIL)
Where not specified, the strain is het-C. Duplications from translocation NM149 include
both the het-c locus and the het-6 locus. Whether het-6 heterozygosity contributes to an
incompatible phenotype detected using NM149 can be determined by progeny-testing with
AR18 or P2869.
het-6PA (Panama CZ30.6, CZ30.4 (het-C?)) 1131 1130
het-6PA (Probably het-C) 2189 2190
het-6PA arg-12 (b9 from Spurger P836) 7350 7351
T(IIL VR)NM149 het-6PA (b7 from P836) 7352 7353
T(IIL VR)NM149 het-6PA (Probably het-C) 2647 2188
het-6OR (OR wild types) 2489 4200
un-24 het-6OR 7354
T(IIL IIIR)AR18 het-6OR 1561 1562
T(IIL VI)P2869 het-6OR 1828 1829
T(IIL VR)NM149 het-6OR 3879 3880
T(IIL VR)NM149 het-6OR (het-c) 1483 1482
T(IIL VR)NM149 het-6OR pyr-4 3136
T(IIL VR)NM149 het-6OR ro-3 2011 2012
het-7 (IIIR)
het-7LI (Liberia UA-1) 961
het-7OR (OR wild types) 2489 4200
T(IIIR X;IIIR;VIIL)D305 het-7OR 2139 2140
T(IIIR X;IIIR;VIIL)D305 het-7OR dow 3150 3151
het-8 (VIL)
het-8PA (Panama CZ30.6, Marrero-1d) 1131 2224
T(VIL IR)T39M777 het-8PA 7413 7412
het-8OR (OR wild types) 2489 4200
chol-2 nit-6 het-8OR 7212
ser-6 het-8OR ad-8 7213
T(VIL IR)T39M777 het-8OR 2133 2134
FGSC No.
Genotype A a
T(VIL IR)T39M777 nit-6 het-8OR 7409 7408
T(VIL IR)T39M777 ser-6 het-8OR 7406 7407
T(VIL IR)T39M777 ad-8 het-8OR 3187 3188
het-8HO (Houma-1n, 1 ) 2220 3943
chol-2 nit-6 ser-6 het-8HO 7485 7486
T(VIL IR)T39M777 het-8HO 7411
het-9 (VIR)
het-9PA (Panama CZ30.6) 1131
het-9OR (OR wild types) 2489 4200
T(VIR IVR)AR209 het-9OR 1931 1932
het-10 (VIIR)
het-10CR (Costa Rica UFC205a) 851
het-10OR (OR wild types) 2489 4200
T(VIIR IL)5936 het-10OR 2104 2105
mating type (IL)
(In am1, the mating and het-incompatibility functions of a are both inactive;
in am33, the hetfunction is inactive but the a mating function remains intact.
(Griffiths and DeLange 1978 Genetics 88:239-254).
tol is an unlinked recessive suppressor of A/a het-incompatibility.)
am1 ad-3B cyh-1 4564
am33 5382
am33 arg-3 5383
am33 ad-3B 4568
tol (N83) 2338 1946
tol trp-4 2336 2337
leu-3 suc; tol pan-1 7322
leu-3 cyt-1 arg-3; tol 7337
T(IL=> IIR)39311 1245 1246
T(IL=> IIR)39311 am33 6705
T(IL=> IIR)39311; tol trp-4 2985 2976
T(IL=> IIR)39311 ser-3 arg-1; tol 3220
In(IL=> IR)H4250 1563 1564
In(IL=> IR)H4250; tol 1947 2975
In(IL=> IR)H4250 leu-3; tol 3253 3254
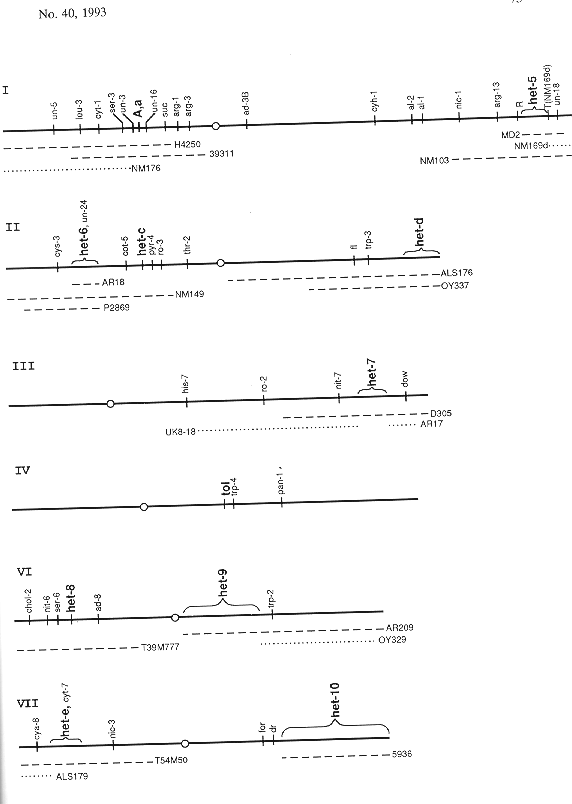
Figure 1. N. crassa linkage groups showing the sequence of markers and rearrangements relevant to known het loci. Dashed lines below the linkage groups show the extent of duplications produced from the crosses between normal sequence and the respective chromosome rearrangements that produce duplications containing a het locus. Dotted lines below the maps show the extent of duplications that do not include a known het locus. For example, in a cross of insertional translocation AR18 het-6OR × normal sequence het-6PA, one third of the viable progeny are duplicated for the segment marked AR18. These duplications are heterozygous het-6OR/het-6PA but haploid for genes outside the duplication. For more complete maps, see Fungal Genet. Newsl. 39:61-70, 1992 or Genetic Maps, 6th edition.