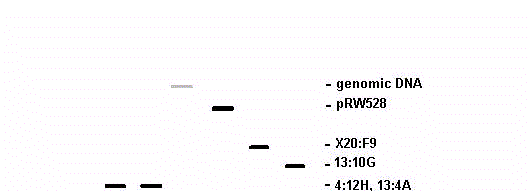
Fig. 1. Hybridization of labeled rDNA from plasmid pRW528 to vacuum-blotted cosmid and genomic DNAs. The membrane shown contained the cosmid DNAs and 35 additional non-hybridizing cosmid DNAs.
In our initial experiments, colony filters from the Orbach-Sachs and Vollmer-Yanofsky libraries were probed with labeled, subtracted cDNAs. Previously identified rDNA cosmids from the Vollmer-Yanofsky library all produced strong signals on the autoradiograms. Only one cosmid from the Orbach-Sachs library, X20:F9, produced a comparable signal.
More definitively, we probed the Orbach-Sachs library for rDNAs using sequences from pRW528, a pBR322-derived plasmid that contains the N. crassa 17S and 26S rRNA genes (Russell et al. 1984 Mol. Gen. Genet. 196:275-282; Butler and Metzenberg 1989 Genetics 122:783-791). An 8.7 kbp BamHI fragment carrying the rRNA genes from this plasmid was gel purified and labeled with 32P. This fragment was used to probe blotted colonies in the Orbach-Sachs library (with the exception of those on plate X25). This screen again revealed only X20:F9 as a strongly hybridizing cosmid.
In a final experiment, we isolated DNA from cosmid X20:F9 and 88 other Orbach-Sachs cosmids, which included every possible candidate for an above-background hybridization to pRW528 or our labeled cDNAs. These DNAs were bound to nylon membranes (Zetabind) using a Bio-Rad Bio-Dot SF microfiltration apparatus and were probed with the labeled pRW528 rDNA fragment. We also included in these experiments cosmid DNA preparations from 21 strains from the Vollmer-Yanofsky library that had hybridized weakly or strongly to labeled cDNAs, and which did not overlap with the previously identified rDNA cosmids from this library. The 110 cosmid DNAs in question were distributed across three separate membranes. We included on each membrane cosmid X20:F9 and three control DNAs: genomic N. crassa, pRW528, and cosmid 13:10G (a known rDNA cosmid from the Vollmer-Yanofsky library).
Restriction digests and gel analyses were used to insure that DNA loading was approximately equal for all cosmids. Hybridization to the three controls was strong on all autoradiograms. Among Orbach-Sachs cosmids, there was detectable hybridization to only X20:F9, which was consistently strong. Three cosmids (in addition to 13:10G) from the Vollmer-Yanofsky library showed strong hybridization: 25:10D, 4:12H and 13:4A. We suggest that these three Vollmer-Yanofsky cosmids contain rDNA. A representative hybridization is shown in Fig. 1.

Fig. 1. Hybridization of labeled rDNA from plasmid pRW528 to
vacuum-blotted cosmid and genomic DNAs. The membrane shown
contained the cosmid DNAs and 35 additional non-hybridizing
cosmid DNAs.
We are unable to explain the nearly total lack of rDNA inserts in the Orbach-Sachs cosmid library. In N. crassa the rRNA genes are tandomly arranged in approximately 9.2 kbp units (Russell et al. 1984 Mol. Gen. Genet. 196:275-282). The average cosmid therefore would be expected to contain four to five rDNA repeats. One possibility is that promoters in the pMOcosX vector cause significant transcription of N. crassa rRNA genes in host cells, resulting in inhibition of bacterial growth. If this is the case, it should perhaps provide a cautionary note to users of the library who are interested in other genes.