A method for the preparation of intact chromosomal DNA of Pycnoporus cinnabarinus
Cifuentes, V., C. Martinez and G. Pincheira Laboratorio de Genetica, Dpto. de Ciencias Ecologicas,
Facultad de Ciencias, Universidad de Chile, Casilla 653, Santiago, Chile.
We describe an improved procedure to obtain chromosomal DNA of Pycnoporus cinnabarinus, which
was used in pulse field gel electrophoresis employing a contour-clamped homogenous electric field
(CHEF). Four bands of chromosomal DNA were separated. The procedure may be useful to study the
genome of other filamentous fungi.
P. cinnabarinus is a basidiomycete capable of degrading components of lignocellulose in nature. Due
to the difficulties of conducting classical genetic experiments in this white rot fungus, its genome is totally
unknown. Therefore it is interesting to study its DNA through pulse field gel electrophoresis.
The P. cinnabarinus strain Pc58, isolated from carpophores collected in the rain forest of southern
Chile, was grown for 8 days at 30oC in solid PC medium (Vogel's medium, with a concentration of
nitrogen equal to 1/3 of the standard composition, supplemented with 0.2% malt extract, 10 g/ml
thiamine, 0.4% glucose and 2% agar-agar).
Intact chromosomal DNA of P. cinnabarinus strain Pc58
was prepared by a method based on embedded agarose
(Schwartz et al. 1984 Cell 37:67-75). 10(10) spores were
suspended in 40 ml of PC medium. They were pelleted at
5000 rpm in a SW40 rotor (Sorvall) for 10 min at 4°C,
washed twice with the same medium and germinated for 4
h at 30°C in a rotary shaker at 120 rpm. The germinated
spores were pelleted at 5000 rpm in a SW40 rotor for 10
min at 4°C and washed twice with 50 mM EDTA pH 7.5,
then suspended in 500 µl of 50 mM EDTA pH 7.5. After
addition of 75 µl of Zymolase 100T (Seikagaku Kogyo Co.,
Tokyo, 10 mg per ml 10 mM sodium phosphate containing
50% glycerol) and 100 µl of Novozyme 234 (20 mg per ml
50 mM EDTA pH 7.5), cells were incubated at 37°C with
1.5 ml of 1% low-gelling-temperature agarose (in 125 mM
EDTA pH 7.5), prewarmed to 50°C and then cooled to
40°C. The suspension was incubated for 30 min at 37°C,
distributed in 100 µl molds (LKB cat. 2015-404), and
allowed to solidify for 30 min at 4°C. The agarose inserts
were added to 4 ml of LET buffer (0.5 M EDTA, 10 mM
Tris pH 7.5, 7.5% 2-mercaptoethanol) plus 200 µl of
Zymolase 100T (10 mg/ml) and 200 µl Novozyme 234 (20
mg/ml) and incubated for 18 h at 37°C. The inserts were
incubated in NDA (10 mM Tris, 0.5 EDTA, 1% lauroyl
sarcosine [pH 9.5]) plus 2 mg/ml of proteinase K for 24 h
at 50°C. This solution plus proteinase K was replaced once
and incubated overnight at 50°C. Inserts were washed twice
with 50 mM EDTA pH 7.5 and stored in 50 mM EDTA pH
7.5 at 4°C.
CHEF gel electrophoresis was performed in a LKB pulsaphor, using a hexagonal electrode (LKB cat.
2015-203) and 0.8% agarose gels in 0.1 M TBE. The voltage used, switching pulse and the running times
of electrophoresis are described in Fig. 1. Gels were stained with ethidium bromide (1 µg/ml for 60 min
and destained in distilled water for 10 min. Intact chromosomal DNA of Saccharomyces cerevisiae strain
AB 1380 was used as a standard.
The results of electrophoresis showed four bands of chromosomal DNA (Fig. 1). Densitometric
analyses suggest that one of them migrated as a single band, another migrated as a doublet, and the other
two bands migrated as a triplet and a quadruplet respectively. The method described here for preparing
DNA from P. cinnabarinus and its analyses by using pulse fields gel electrophoresis is an alternative way
to karyotyping this fungus. In addition, it should be useful for studying other filamentous fungal species.
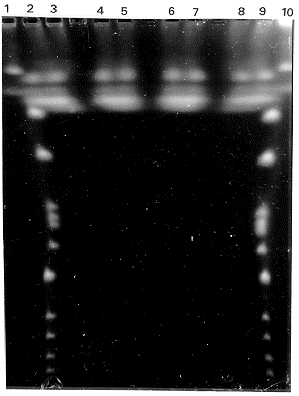
Figure 1. Separation of P. cinnabarinus intact chromosomal DNA on a CHEF gel. Agarose gel was
electrophoresed at 13°C with three pulse intervals of 60 sec, 90 sec and 120 sec with a duration of 15 hr,
7 hr and 20 hr with voltages of 200 V, 200 V and 150 V respectively. Lanes 1 and 10 correspond to S.
cerevisiae strain AB1380; lanes 2 to 9 correspond to P. cinnabarinus strain Pc58: lanes 2, 3, 4 and 5
(preparation 1); lanes 6, 7, 8 and 9 (preparation 2).
Return to the FGN 37 index
Go to the FGSC main page
