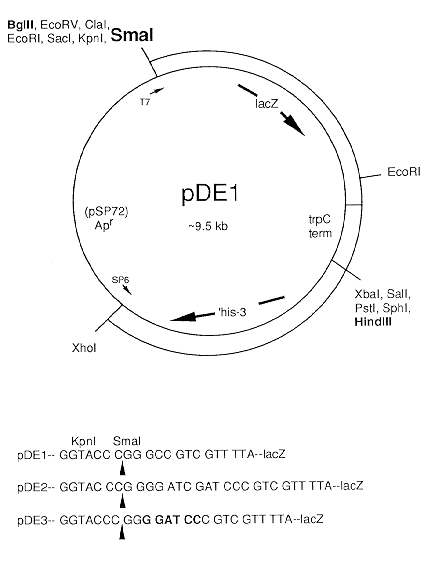
The use of lacZ as a reporter gene requires appropriate plasmid constructions to produce fusions of fungal promoter and translations initiation sequences to the E. coli lacZ gene. I have constructed three plasmids which may facilitate construction of lacZ fusions.
The three plasmids are identical except for a BamHI site adjacent to a unique SmaI site in three different reading frames with respect to lacZ. Thus any blunt-ended fragment containing a functional promoter and translation start site can be used to produce an in frame translational fusion by choosing the appropriate plasmid vector.
The vector contains E. coli lacZ from pMC1871 (Shapira et al. 1983, Gene 25:71-82) followed by the terminator of Aspergillus nidulans trpC obtained from plasmid DH25 (Cullen et al. 1987, Gene 57:21-26). It also contains the truncated his-3 gene from pH303 (see accompanying paper) which allows targeted integration of the lacZ fusions at the his-3 locus in FGSC strain 462. The vector for these plasmids is pSP72 (available from Promega Corporation, Madison, WI).
Figure 1 shows a map of pDE1. The plasmid is approximately 9.5 kb in size. The vector sequence of pSP72 runs from the XhoI site to the SmaI site in all three plasmids. The BamHI site was used to clone the BamHI to XbaI fragment from pDH25 containing the trpC terminator. The HindIII to SmaI fragment containing the his-3 gene segment from pH303 was cloned into the pSP72 polylinker at HindIII to PvuII. Finally the BamHI fragment containing lacZ was cloned into the BamHI site adjacent to the SmaI site of the pSP72 polylinker. The BamHI site at the lacZ/trpC terminator junction was destroyed by cleavage with BamHI and filling of the 5' overhangs followed by ligation. Plasmids pDE1, pDE2 and pDE3 differ at the BamH1 site adjacent to the SmaI site. The plasmid with this BamH1 site left intact was designated pDE3. pDE3 was digested with BamHI and ligated following i) mung bean nuclease digestion to generate pDE1 or ii) Klenow fragment of DNA polymerase and dNTP's to generate pDE2.
The nucleotide sequence of this region was determined on both strands to verify the sequence shown in Fig. 1 for each plasmid. The arrows show the cleavage point of SmaI. The codon triplets of lacZ following the SmaI site are also shown. The unique sites in the plasmids are shown in bold: BglII, SmaI and HindIII. pDE3 also contains the unique BamH1 site adjacent to SmaI.
The lacZ/trpC terminator can be removed as a cassette with BglII (or SmaI) and HindIII for transfer to new plasmids (for example, a plasmid conferring benomyl resistance). Another feature of the plasmid is the T7 and SP6 promoters for synthesizing RNA from each direction for making probes for Southern analysis. The "T7" sequencing primer and lacZ "-40" primers can be used for sequencing fragments cloned into the plasmids for verification of constructions.
The lacZ gene of pDE3 is in frame with an upstream ATG in the vector sequence and potentially could be used to make transcriptional fusions from promoters cloned into the BglII site; however, the ATG has a poor sequence context for translation in N. crassa.
The possible limitation of using these plasmids in the low transformation frequencies obtained by requiring integration at the his-3 locus in FGSC strain 462. These plasmids have been deposited with the Fungal Genetics Stock Center. This work was performed in the laboratory of Dr. Charles Yanofsky.
