A simple procedure for the isolation of fungal DNA for dot blot analysis
R. Geisen - Institute of Microbiology, Toxicology and Histology, Federal Centre for Meat Research, 8650
Kulmbach, Federal Republic of Germany
A rapid method for the detection of transforming sequences in a fungal strain would be advantageous
when trying to determine if unselected sequences are present. Colony hybridization protocols for
filamentous fungi have been developed (Stohl and Lambowitz, 1983. Anal. Biochem. 134:82-85; Paietta
and Marzluf, 1984. Neurospora Newsletter 31:40) and a modified method thereof was described (McClung
and Dunlap, 1988. Fungal Genetics Newsletter 35:26-27). In this report a simple method for the isolation
of fungal DNA from a single transformed colony suitable for dot blot analysis is described. If the original
transformant colony is not too small it is sufficient to extract the DNA of one half of that colony. That
means that no subculturing is necessary and the results can be available within 24 h starting from the
transformant colony.
1. Transfer one half of the transformant colony (or approximately 0.1 g of mycelium from a liquid culture)
into a microfuge tube.
2. Add 1 ml of lysis buffer (50 mM EDTA, 0.2% SDS) and 0.1 g of Alumina (Type A5, Sigma).
3. Mix in a microfuge tube mixer (for this protocol an Eppendorf micro tube mixer was used) for 30 min.
4. Centrifuge at 5000 rpm for 5 min.
5. Transfer the supernatant (750 µl) to a new microfuge tube.
6. Extract once with phenol, phenol/chloroform and chloroform each.
7. Add 2 M sodium acetate to a final concentration of 0.2 M.
8. Precipitate the DNA with an equal volume of isopropanol.
9. Dry the pellet and redissolve in an appropriate volume of TE buffer.
With this DNA isolation procedure a contransformation experiment using Penicillium nalgiovense
ATCC 66742 as host organism was checked by dot blot analysis. The vector p3SR2 which carries the
amdS gene (Hynes et al. 1983. Mol. Cell. Biol. 3:1430-1439) as a marker was used as a selectable plasmid.
As a nonselectable plasmid, pELN5-lac was used. pELN5-lac carries the E. coli ß-galactosidase gene
under the control of the promoter of the oliC31 gene (Ward and Turner 1986. Mol. Gen. Genet.
205:331-338) and the terminator of the trpC gene (Mullaney et al. 1985. Mol. Gen. Genet. 199:37-45)
from Aspergillus nidulans.
One microgram of each of the two plasmids were cotransformed and positive transformants were
selected for the presence of the amdS marker by growth on acetamide minimal medium. The DNA of
one half of the resulting transformant colonies (diameter approximately 1 cm) was isolated as described
above. Under these conditions 0.1-0.5 µg of DNA was isolated. The whole amount of DNA was used for
dot blot analysis. Hybridization was carried out using a digoxygenin labelled DNA fragment which carries
the E. coli ß-galactosidase gene. Figure 1 shows the result of this dot blot analysis. The amount of DNA
isolated from a single colony gives clear positive signals. In this experiment a contransformation
frequency of 70% was reached. After initial screening with the described method, single spores of the
cotransformants were subcultured on acetamide minimal containing 0.1 µg/ml X-gal (5-bromo-4-chloro-3-
dinolyl-ß-D-galactopyranoside). All strains with positive dot blot signals showed, in contrast to the
untransformed strain, a blue color of the mycelium indicating the presence of the E. coli ß-galactosidase
gene. These results suggest that the cotransformed DNA which was detected by the described method
was not in an abortive stage.
The advantage of the described method is the fact, in contrast to the method of McClung and Dunlap
(1988. Fungal Genetics Newsletter 35:26-27), which starts from cultures on agar slants as source material,
that the original transformed colony can be analyzed for the presence of specific DNA sequences. There
is no need for the preparation of protoplasts and the background due to unspecifically bound hybridization
probe sequences in very low.
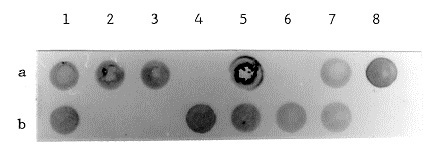
Figure 1. Dot blot analysis of the DNA of Penicillium nalgiovense cotransformants.
The DNA of 14 cotransformants of Penicillium nalgiovense ATCC 66742 transformed with equimolar
amounts ( 1 µg each) of p3SR2 and pELN5-lac was isolated as described in the text. The DNA was
resuspended in 100 µl TE, heated at 95°C for 10 min and cooled immediately on ice. The DNA was
transferred to nitrocellulose filters (Schleicher & Schull, BA 83) using a Schleicher and Schull Minifold
I filtration device. The filter was baked for 2 h at 80°C and prehybridized for 2 h at 68°C. A 3.0 kb DNA
fragment carrying a part of the E. coli lacZ gene was digoxygenin labelled as described by the
manufacturer (Boehringer, Mannheim) and used as a hybridization probe. Hybridization was carried out
at 68°C overnight. The washing and developing procedure was performed as described by the
manufacturer. The dots 1-7, a+b represent the analyzed samples. As a positive control (8a) 10 ng of
pELN5-lac and as a negative control (8b) the DNA of the untransformed strain was used.
Return to the FGN 37 index
Go to the FGSC main page
