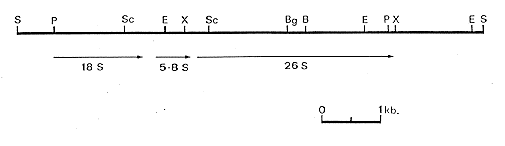
Figure 1. Restriction endonuclease map of the A. niger ribosomal repeat. Restriction endonuclease sites: S - SalI; P - PstI; Sc - SacI; E - EcoRI; X - XhoI; Bg - BglII; B - BamH1 The lines below the map indicate the positions of the rRNA genes.
The ribosomal RNA genes have been used in studies of a variety of phenomena. These include studies of molecular phylogeny, transcription, recombination and transformation. In Saccharomyces cerevisiae, where integrative transformation is achieved by homologous recombination, the presence of the ribosomal repeat unit in the transformation vector greatly increases the frequency of transformation (Szostak and Wu 1979. Plasmid 2:536- 554; Smolik-Utlaut and Petes 1983. Mol. Cell. Biol. 3:1204-1211). In the filamentous fungus Aspergillus nidulans, where integrative transformation can be achieved either via homologous or heterologous recombination, the presence of ribosomal repeat sequences in the transformation vector has no effect on the frequency of transformation (Tilburn et al. 1983. Gene 26:205-221). [Ed.: This is also the case in Neurospora crassa: Russell et al. 1989. BBA 1008:243-246]. Here we report the molecular cloning of the ribosomal repeat unit from Aspergillus niger. We have found that the presence of cloned ribosomal DNA sequences from A. niger increases the frequency of transformation in A. niger.
1. Cloning of the ribosomal repeat unit of A. niger
A genomic library of wild type A. niger (Kelly and Hynes 1985. EMBO J 4:475-479)
DNA, partially digested with MboI, was constructed in the lambda based replacement vector
EMBL 3. The library was screened with a 32P labelled plasmid probe, pMN1, containing
the A. nidulans ribosomal repeat unit (Borsuk et al. 1982. Gene 17:147-152). Several
plaques were identified that hybridized to pMN1. SalI digested DNA prepared from these
plaques showed a band of approximately 7.8 kb, and Southern blot analysis showed this
band to have homology to pMN1. That this band represented an entire ribosomal repeat
unit was inferred from the finding from Southern blot analysis that A. niger DNA cut with
a series of enzymes, including SalI, BamHI and BglII, gave a hybridizing band of
approximately 7.8 kb representing one ribosomal repeat unit. This 7.8 kb sequence was
cloned into the SalI site of pBR322 and the recombinant plasmid was designated pANiR1.
A partial restriction map was determined and is shown in Figure 1. The coding regions are
also shown as determined both by hybridizing restriction fragments of pANiR1 to Northern
blots of total RNA extracted from a wild type strain of A. niger and by hybridizing
ribosomal RNA probes to Southern blots of various digests of pANiR1. The position of the
genes, and of restriction sites, was compared with those found by Borsuk et al. (1982. Gene
17:147-152) and Lockington et al. (1982. Gene 20:135-137) in the cloned A. nidulans
ribosomal repeat DNA. The position of the genes was found to be conserved, as were the
restriction sites within the coding regions. The restriction sites within the non-transcribed
spacer region were not conserved.

Figure 1. Restriction endonuclease map of the A. niger ribosomal repeat. Restriction endonuclease sites:
S - SalI; P - PstI; Sc - SacI; E - EcoRI; X - XhoI; Bg - BglII; B - BamH1
The lines below the map indicate the positions of the rRNA genes.
2. The effect of the ribosomal repeat unit of A. niger of the frequency of transformation
Transformations of A. niger were performed using the amdS gene of A. nidulans as a
dominant selective marker (Kelly and Hynes 1985. EMBO J 4:475-479). The plasmid used,
p3SR2, contained the amdS gene cloned into pBR322 (Hynes et al. 1983. Mol. Cell. Biol.
3:1430-1439). Transformation frequencies were compared in experiments using p3SR2
alone, p3SR2 plus equimolar amounts of pMN1, and p3SR2 plus equimolar amounts of
pANiR1 (Table I). Higher frequencies of transformation were consistently found when
pANiR1 was present, but not when pMN1 was present. This result was repeatable, and did
not depend on the particular amdS containing plasmid used. Transformation frequencies
of A. niger using amdS selection were low, possibly due to the observation that
transformants contain multiple copies of the amdS gene, and a transformant containing a
single copy of the amdS gene may not grow sufficiently to be detected.
Table I: Transformation of A. niger in the presence of the cloned ribosomal repeat from A. niger and A. nidulans
Number of amdS+ transformants per microgram DNA
Transforming DNA Experiment 1 Experiment 2
p3SR2 15 26
p3SR2 + pMN1 24 30
p3SR2 + pANiR1 167 138
Results shown are of two independent experiments, each with 2 x 10(4) viable
protoplasts per treatment. In six experiments, the average fold increase was
five.DNA was extracted from eight strains cotransformed with pANiR1 and p3SR2, digested with HindIII, separated by agarose gel electrophoresis, transferred to nylon membrane, and hybridized with the insert of p3SR2. The membrane was washed and rehybridized with the insert of pANiR1. There are no HindIII sites in the ribosomal repeat unit, and as expected, the insert of pANiR1 hybridized to a band of very high molecular weight, too large to quantify on standard agarose gels. There are no HindIII sites in p3SR2, and no hybridization is observed between p3SR2 and wildtype A. niger DNA. In the transformant DNA, the amdS probe also hybridized to a band of very high molecular weight. Thus it is probable that the amdS sequences integrated at the ribosomal repeat region.
In order to determine if this result was entirely dependent upon the selectable marker in the recipient strain, transformations were performed using the cloned A. niger pyrG gene (pAB4.1) as a selectable marker in a pyrG auxotrophic strain of A. niger (van Hartingsveldt et al. 1987. Molec. Gen. Genet. 206:71-75). The plasmid pAB4.1 transformed the A. niger strain AB4.1 at a much higher frequency than observed for p3SR2 and the wild type strain. Numbers of transformants in experiments using pAB4.1 alone were compared with numbers of transformants using both pAB4.1 and pANiR1. In four independent experiments, the transformation frequency was 3.7-, 3.9-, 4.0- and 4.5-fold higher in the presence of pANiR1 than in its absence. These experiments showed that the effect of pANiR1 on transformation frequency was neither recipient strain nor selectable marker dependent. Various subclones of pANiR1 were tested in an attempt to localize the effect. Neither the EcoRI-EcoRI fragment nor the small EcoRI-SalI fragment had an effect on transformation frequency when present as inserts in pBR322 in cotransformation experiments. Further, the increase in transformation frequency was not apparent when the ribosomal repeat unit was inserted into the same plasmid as the amdS selectable marker, but this may be due to the reduction in transformation frequency due to the very large size of the plasmid masking the five fold effect.