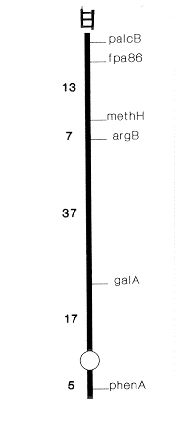
Figure 1. Relevant parts of linkage group III of A. nidulans showing the location of fpa86 with respect to other markers
A number of different mechanisms for p-fluorophenylalanine (FPA) resistance have been described in Aspergillus nidulans. We present data on a new locus for FPA resistance and its biochemical change. Before this report, 20 FPA resistant loci conferring resistance to this amino acid analogue have been identified by us (Tiwary et al. 1987 Curr. Microbiol. 15:305-311) and two by Kinghorn and Pateman (1975 J. Gen. Microbiol. 86:174-184). These mutations have been mapped on five out of eight linkage groups of A. nidulans, there being no report of any mapping of FPA resistant loci on linkage groups III,IV or VII. A new class of FPA resistant mutants exhibiting a reduced level of phenylalanyl-tRNA synthetase was identified in a selection scheme using nitrate as the nitrogen source (Tiwary et al. 1987 Mol. Gen. Genet. 209:164-169). We have slightly modified the technique for the selection of analogue resistant mutants by substituting aspartic acid (25 mM) for sodium nitrate (0.6%) as the nitrogen source in the Czapek-Dox medium in the presence of 30 mg/ml FPA. A higher rate of survival was found on medium containing aspartate than nitrate.

Figure 1. Relevant parts of linkage group III of A. nidulans showing the location of fpa86
with respect to other markers
Nine mutants selected on an aspartate medium by using biA1;phenA3 (Glasgow stock no. 310) as a starter strain were found to be allelic and were given the isolation number fpa86. The LD50 value of FPA for the mutant was found to be 35 mg/ml whereas it was 12 mg/ml for the starter strain. Dominance test in the heterozygous diploid (fpa86/MSF) revealed that the new mutation was recessive to its wild-type allele. This mutation was located on linkage group III by mitotic analysis of segregants obtained after haploidization on chloral hydrate (0.02 M) of the heterozygous diploid synthesized between fpa86 and MSF of McCully and Forbes (1965 Genet. Res. 6:352-359). With the help of suitable crosses (Table I), fpa86 was mapped on the left arm of linkage group III, about 13 map units left of the methH locus (Fig. 1). We have also undertaken biochemical characterization of this new mutant. Table II indicates that in affinity chromatographic purification, the enzyme Phe-tRNA synthetase from the wild type as well as the mutant strain reveals similar affinity for the analogue (used as ligand) in the column. However, when the analogue (1 mM) was used as substrate in a catalytic reaction, the wild-type enzyme shows a much higher affinity for the analogue (measured in terms of ppi released) as compared to the mutant enzyme which catalyzes a very low ppi generation due to the slower rate of ATP-amino acid (analogue) exchange (Table III). This phenomenon of slow analogue activation by the mutant enzyme during the process of translation perhaps leads to its slow incorporation into proteins ultimately bringing about resistance to the analogue.
Phe-tRNA synthetase of A. nidulans has bee reported to be a quaternary structure (alpha2ß2), the molecular mass of the alpha-subunit being 67 kdal and that of the ß-subunit 75 kdal (Rauhut et al. J. Biol. Chem. 259:6370-6375). From the results obtained, it appeared most likely that a mutation in fpa86 caused conformational change in one of the subunits. To test the validity of this assumption, SDS-polyacrylamide gel electrophoresis (7.5%) of Phe-tRNA synthetase (Fig. 2) purified from the active fractions of the column (both wild type and the mutant) was carried out (Laemmli 1970 Nature 227:680-685). Two bands corresponding to 66 and 59 kdal were clearly visible in the case of the wild type enzyme, whereas the mutant synthetase showed the appearance of an additional polypeptide band migrating slightly faster than the ß-subunit.
This new band could be considered as a modified ß- subunit which results in functional inactivity of the enzyme in the mutant strain. However, we have no positive explanation for the discrepancy in observed sizes of the two bands if we compare our result with the earlier report on the molecular mass of the alpha and the ß subunits. Moreover, whether modification in the ß subunit occurs during activation or loading onto tRNAphe of the analogue by Phe-tRNA synthetase is yet to be determined. Experiments to investigate this issue are in progress.
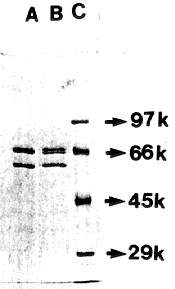
Figure 2. SDS-polyacrylamide gel electrophoresis (7.5%) of Phe-tRNA synthetase. (A)
Wild type (B) fpa86 (C) SDS-high molecular weight protein markers (Sigma).
Chemical modification of Phe-tRNA synthetase in FPA resistant mutants leading to alteration in the binding sites and reduced ability to bind FPA has been reported in E. coli (Fangman and Neidhardt 1967 J. Biol. Chem. 239:1839-1843) and A. nidulans (Tiwary et al. 1987 Mol. Gen. Genet. 209:167-169). It seems most likely that as a consequence of modification in the ß-subunit, there occurs a drastic decrease in the turnover number of the enzyme for the analogue due to lack of anticooperativity between domains. However, in the column the affinity of Phe-tRNA synthetase for the analogue appears to be independent of its turnover number. Furthermore, it does not really appear to matter whether all four sites in the enzyme are active or only one of them is so, because in either case the mutant enzyme is expected to show a similar affinity for the analogue to that of the wild-type enzyme in the column.
Table I. Linkage of fpa86 with other markers of linkage group III
Cross Genotype Markers Genotype of progeny* Recomb.
of strains Considered + + + - - + - - Freq (%)
1. biA1;phenA3 yA2-biA1 S.A. S.A. 127(P) 9(R) 6.61 ± 0.18
fpa86 X galA1-phenA3 S.A. S.A. 141(P) 39(R) 21.66 ± 1.04
MSF galA1-fpa86 S.A. S.A. 51(P) 46(R) 47.42 ± 1.28
phenA3-fpa86 S.A. S.A. 87(R) 82(P) >50
2. biA1;phenA3 fpa86-methH2 17(R) 116(P) S.A. S.A. 12.78 ± 1.07
fpa86 X yA2; fpa86-argB2 28(R) 105(P) S.A. S.A. 21.05 ± 1.64
methH2 argB2 fpa86-phenA3 64(P) 69(R) S.A. S.A. >50
methH2-argB2 81(P) 10(R) S.A. S.A. 10.98 ± 0.67
MSF = suA1adE20 yA2 adE20;acrA1;galA1;pyroA4;facA303;sB3;nicB8;riboB2
* P = Parentals, R = Recombinants, S.A. = Selected againstTable II. Amount of Phe-tRNA synthetase bound to FPA (as ligand) in different fractions
Fraction no. Amount of bound protein eluted
(3 ml each) from the column (in µg/ml)
wild type mutant
1 59.0 57.0
2 58.0 60.0
3 60.5 53.0
4 16.0 20.0
5 7.5 8.0
6 6.0 6.0
7 5.0 4.0
Table III. Amount of ppi released
(in µM) in ATP-amino acid exchange
by wild-type and mutant Phe-tRNA
synthetase.
Fraction no.
(0.2 ml each) Wild type Mutant
1 138 13
2 131 15
3 145 13
4 37 4
5 18 N.D.*
6 18 N.D.
7 11 N.D.