The bli regulon - a network of blue light inducible genes of N. crassa
N.N. Pandit and V.E.A. Russo - Max-Planck-Institut fur molekulare Genetik, IhnestraŠe 73,
D-1000 Berlin 33, Germany.
Several physiological responses of N. crassa are observed when this fungus is exposed
to blue light. Here, we do not intend to make a comprehensive list of all the light effects
observed so far in N. crassa (for a review, see Degli Innocenti and Russo 1984. In "Blue
Light Effects in Biological Systems" ed. H. Senger, Springer-Verlag. pp 213-219.), but point
out only the underlying themes. First, the time interval between the light stimulus and the
observed response can be very different, and ranges from a few minutes to several hours -
or even days - depending on the nature of the physiological response in question. For
instance, alterations in the membrane potential and the synthesis of carotenoids take place
within a matter of a few minutes, whereas an increase in the number of protoperithecia in
an agar plate culture, grown in conditions which favor protoperithecia production, takes 24
hours! Perithecia, the bodies containing products of the sexual cycle, if exposed to light
during early stages of their formation, display phototropism of the perithecial beaks, a
process which takes as many as 12 days.
A second important point to note besides the time element is that the metabolic
stages of the N. crassa life cycle at which the tissues are exposed to blue light, and even the
types of tissues exposed, are different in the physiological responses mentioned above. Thus,
it appears that N. crassa has a genetic apparatus which can be activated by light virtually at
any stage during its life cycle. We shall refer to this genetic apparatus as the bli global
regulon, a conceptual equivalent of the global regulons which have been described in
prokaryotic systems (Hoops and McClure 1987. In "Escherichia coli and Salmonella
typhimurium" ed. F.C. Neidhardt et al. ASM Publications pp 1231-1240). The term "bli"
implies "blue light inducible" to indicate the fact that in cases where the action spectrum was
analyzed, only the blue part of light was observed to be effective.
Lastly, the third important point is that the bli regulon consists of a very large
number of genes. A variety of translatable mRNA species appear in a temporal fashion
only in the mycelia irradiated with blue light. It has been calculated that at least 60 genes
are induced within 30 minutes in mycelia exposed to blue light. This massive gene action
is blocked by the wc-1 or wc-2 mutations (Chambers et al. 1985. EMBO J. 4:3649-3653,
Nawrath and Russo 1990. J. Photochem. Photobiol. 4:261-271). The cDNA clones of four
blue light inducible (bli) genes bli-3, bli-4, bli-7, and bli-13 whose functions are not yet
known, have been obtained. These bli genes define three different classes of genes based
on the time taken for the accumulation of the transcripts at the "steady state level". bli-3
and bli-7 have been shown to have different DNA sequences (Eberle et al., in preparation).
The lag periods observed were 2, 15 and 45 minutes respectively (Sommer et al. 1989.
Nucleic Acids Res. 17:5713-5723).
The groups of genes which have been identified in other ways but are known to be
regulated in response to light include al-1 (Schmidhauser et al. 1990 Mol. Cell. Biol.
10:5064-5070), al-2 (Lauter, Schmidhauser, Yanofsky and Russo, unpublished data) and al-3
(Nelson et al. 1989. Mol. Cell. Biol. 9:1271-1276) - the genes involved in carotenoid
biosynthesis and con-5 and con-10 (Lauter and Russo, unpublished data) - the genes induced
during conidiation (Berlin and Yanofsky 1985. Mol. Cell. Biol. 5:849-855).
How are all these light inducible genes organized in the genome and regulated
precisely, in a temporal fashion? Some of the genes, including al-1, al-2, bli-3, bli-4, con-5
and con-10 are transcribed in concert in less than 5 minutes. The wc-1 and wc-2 genes
which map on the linkage groups VII and I respectively, constitute an important part of the
bli regulon and are believed to be the regulatory genes since none of the blue light inducible
responses analyzed so far are observed in the white collar mutants. Action of the wc gene
products in controlling the light inducible genes must be at the transcript level because the
increase in the amount of al-1 (Schmidhauser et al. 1990. Mol. Cell. Biol. 10:5064-5070),
al-2 (Lauter, Schmidhauser, Yanofsky and Russo, unpublished data), and the bli-3, bli-4, bli-
7 and bli-13 (Sommer et al. 1989. Nucleic Acids Res. 17:5713-5723) transcripts after
exposure to light are not observed in the wc mutants. One possible function involved in this
regulation could be protein dephosphorylation, since the wc mutants clearly show altered
potentials to dephosphorylate proteins. This is the only phenotypic criterion which separates
the wc-1 and wc-2 mutants (Lauter and Russo, 1990. J. Photochem. Photobiol. 5:95-103).
To date, genomic locations of eight light inducible genes have been determined in
N. crassa. Out of three albino genes, al-1 and al-2 map on linkage group I while al-3 maps
on linkage group V. Of the three con genes, con-8 maps on linkage group I, while con-5
and con-10 map on linkage group IV. We have determined the map locations of three more
bli genes, bli-3, bli-4 and bli-7. Essentially, the procedure of Restriction Fragment Length
Polymorphism mapping developed by Metzenberg et al. (1984. FGN 31:35-39) was followed
using the strains described therein. The data will be forwarded to R.L. Metzenberg. The
genomic DNA clones of the bli genes constructed in pUC vector (JĀrgen Eberle, Ph.D.
thesis 1990, Free University, Berlin) were used as probes. The genes bli-4 and bli-7 map
at the same position, linked to ccg on LG II while bli-3 maps near con-5 and con-10 on LG
IV (Figure 1).
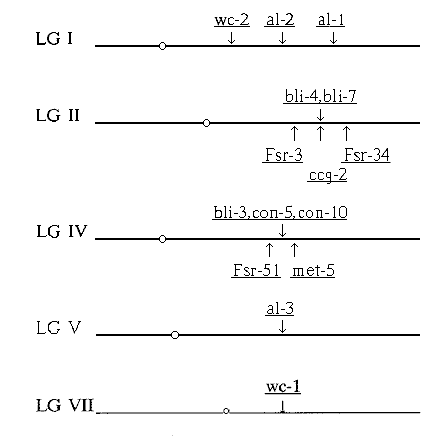
Figure 1. Schematic representation of the linkage groups of N. crassa. Only the relevant linkage groups are shown. Drawn
above the lines are the genes belonging to the bli regulon. For the genes which have been located using the RFLP mapping
technique, flanking genetic markers are shown below the line for comparison.
To summarize, we suggest that the light inducible genes of N. crassa define a global regulon,
termed here as the bli regulon, which is somehow controlled at the transcript level by the wc-1 and
wc-2 gene products. In addition, we have determined the map locations of three newly identified
blue-light inducible genes, and find that most of the light regulated genes are dispersed fairly
randomly on the genome of N. crassa. However, possible existence of small clusters containing bli-4
and bli-7 on linkage group II, and another containing bli-3, con-5 and con-10 on linkage group IV
cannot be ruled out because of the limitations imposed by the available mapping techniques. It is
important to note that the genes which are separated by as many as 50 kb may not always be seen
as separate genes by the RFLP mapping technique.
Acknowledgement: We wish to thank J. Eberle for supplying the E. coli strains harboring the genomic
clones of bli-3, bli-4 and bli-7. We would also like to thank Rudi Lurz for his help in computer work.
