Lipofectin increases the efficiency of DNA-mediated transformation of Neurospora crassa
Claude P. Selitrennikoff(1) and Matthew S. Sachs(2) - (1)Department of Cellular and Structural
Biology, University of Colorado Health Sciences Center, Denver, CO 80262 and
(2)Department of Chemical and Biological Sciences, Oregon Graduate Institute of Science and
Technology, 19600 NW von Neumann Drive, Beaverton, OR 97006.
Transformation of N. crassa using the calcium chloride/polyethylene glycol procedure
(Fincham 1989. Microbiol. Rev. 53:148-170) can yield 100-10,000 transformants per
microgram of input DNA when integration occurs nonhomologously, and considerably lower
numbers of transformants when homologous integration of DNA is required. Increasing the
efficiency of transformation would be useful for screening cosmid libraries and for obtaining
transformants from homologous integration events. In addition, strains that transform
poorly using standard procedures, e.g. the cell-wall-less derivatives of os-1 (Phelps et al. 1990
Curr. Microbiol. 21:233-242) would benefit from improvements in the transformation
procedure. Lipofectin, a liposome preparation formulated from cationic lipids [Bethesda
Research Laboratories, Gaithersburg, MD], has been reported to increase transformation
of the yeast Schizosaccharomyces pombe by 35-fold with plasmids or large (>500 kb) linear
DNA molecules (Allshire 1990 Proc. Natl. Acad. Sci. USA 87:4043-4047). In an effort to
increase the transformation efficiency of N. crassa, we tested the effects of including
lipofectin in a standard transformation procedure.
Germinated macroconidia of wild type N. crassa (74-OR23-1VA) were grown,
harvested and counted. Protoplasts were obtained by treatment with Novozym 234 as
described (Orbach et al. 1986 Mol. Cell. Biol. 6:2452-2461). Protoplasts of the os-1
derivative TM-1 were grown and harvested as described (Phelps et al. 1990 Curr. Microbiol.
21:233-242). Protoplasts of both strains were washed twice with 1 M sorbitol and once with
STC (1 M sorbitol, 50 mM Tris-HCl pH 8.0, 50 mM CaCl2). Protoplasts were resuspended
in STC (0.4 ml per 5 x 10(7) cells). Then 0.1 ml PTC (40% polyethylene glycol 3300 [Sigma
Chemical Co., St. Louis, MO], 50 mM Tris-HCl pH 8.0, 50 mM CaCl2) and 5 ul of dimethyl
sulfoxide were added per 5 x 10(7) cells. Competent protoplasts were stored at -80 C after
aliquoting into eppendorf tubes; aliquots in tubes were transferred directly from wet ice to
the -80 C freezer.
For transformation, cells were thawed on ice. DNA was premixed with heparin (25
ul; 125 ug) in 12 x 75 mm polystyrene tubes [Falcon 2058 tubes; Becton Dickinson Labware,
Lincoln Park, NJ]; cells (0.1 ml) were added and the mixtures incubated on ice for 30 min.
Lipofectin was added, and incubation continued for 15 min at room temperature. One ml
of PTC was added, and after gentle mixing, suspensions were incubated 20 min at room
temperature. Aliquots were added to molten regeneration agar overlayed on minimal plates
containing 1 ug/ml benomyl (DuPont, Wilmington, DE) or 200 ug/ml Hygromycin B
(Boehringer Mannheim, Indianapolis, IN). Plates were scored after 48 h incubation at 34 C
(for wild type) or after 96 h at 25 C (for TM1).
Inclusion of lipofectin at a concentration of 35 ug per ml of wild type protoplasts in
the standard protocol increased yields of transformants by nearly 10-fold (Fig. 1) when
expression of a semi-dominant allele of the tub-2 gene (Orbach et al. 1986 Mol. Cell. Biol.
6:2452-2461), which confers benomyl resistance, was selected. Lipofectin at concentrations
higher than 50 ug/ml did not result in increases in transformation (not shown). Similar
results were obtained using a library of cosmid DNA to transform N. crassa to hygromycin
resistance (Fig. 2). The efficiency of transformation of TM-1 protoplasts to benomyl or
hygromycin resistance, using pMO63 or pMOcosX cosmid library DNA, respectively,
increased 3-4 fold (average of 4 transformations) when lipofectin was included in the
transformation procedure (data not shown). Addition of PTC appeared necessary for
efficient transformation of N. crassa with or without added lipofectin; preincubation of
DNA with lipofectin prior to addition of spheroplasts did not increase transformation
efficiency compared to controls lacking lipofectin. We have limited results concerning the
stability of recovered transformants in that 15/15 colonies recovered from primary
transformation plates were stable (i.e., grew on hygromycin-containing medium for at least
two transfers.
Our results show that addition of lipofectin to a standard N. crassa transformation
protocol increases the number of recovered transformant colonies 3-10 fold. These
increases should prove helpful for experiments in which the highest possible transformation
efficiencies are desired.
Acknowledgements: We thank Dr. N. Raju for processing Fig. 2. CPS thanks Dr. David
Perkins and Dr. Charles Yanofsky for their gracious hospitality while on sabbatical leave at
Stanford University. MSS thanks Dr. Yanofsky for helping to support his postdoctoral study.
This work was supported in part by NIH and NSF grants to CPS.
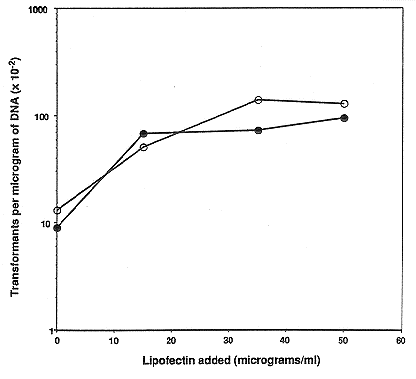
Figure 1: Effects of lipofectin concentration on transformation of wild type N. crassa with
a plasmid conferring benomyl resistance. Wild type protoplasts were incubated with 2 ug
(open symbols) or 5 ug (closed symbols) of pMO63 DNA and 0, 15, 35, or 50 ug/ml of
lipofectin. The number of benomyl-resistant colonies obtained per microgram of DNA at
each lipofectin concentration is plotted as a function of lipofectin concentration. pMO63
contains the mutant tub-2 gene that confers benomyl resistance on a 3.1 kb HindIII
fragment.
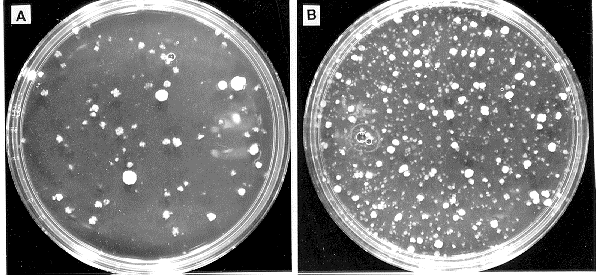
Figure 2: Lipofectin increases the efficiency of transformation of N. crassa to hygromycin resistance with cosmid
DNA. Wild type protoplasts (0.1 ml) were incubated with 1 ug of pooled cosmid DNA in vector pMOcosX and
(A) no lipofectin or (B) 35 ug/ml lipofectin. One-tenth of the transformation mixture was plated on each plate.
The pMOcosX library is available through the FGSC.

