Characterization of a mutation in a strain of Penicillium camembertii affecting the
production of cyclopiazonic acid.
Rolf Geisen - Federal Research Centre for Nutrition, Institute for Hygiene and Toxicology,
Engesserstr. 20, 7500 Karlsruhe, Germany
Penicillium camembertii is a filamentous fungus used for the production of mold-
fermented white cheese. It is a domesticated form of Penicillium commune, especially
adapted to the food environment (Pitt et al. 1986. Food Microbiol. 3:363-371). Despite its
use as food starter culture, P. camembertii is able to produce cyclopiazonic acid (CA), a
secondary metabolite toxic to animals and humans (Holzapfel 1968. Tetrahedron 24:2101-
2119); Le Bars 1979. Appl. Environ. Microbiol. 38:1052-1055). The synthesis of CA starts
from the amino acid tryptophan with the condensation of acetyl-CoA and isoprenoids,
especially dimethyl allylpyrophosphate (Holzapfel 1980. In: P.S. Steyn, The biosynthesis of
mycotoxins, Academic Press). Acetyl-CoA is also the direct precursor of the isoprenoids.
To define the genes responsible for the production of cyclopiazonic acid, P.
camembertii Ps 912 was treated with nitrous acid and screened for CA negative mutants.
One mitotically stable strain was isolated with only 1-2% of CA production compared to the
wild type (Geisen et al. 1990. Appl. Environ. Microbiol. 56:3587-3590).
To localize the mutation either in the pathway for biosynthesis of isoprenoids or in
the biosynthetic branch between tryptophan and CA, radioactive labelling either with 3H-
tryptophan or with 14C-acetate was carried out. For this purpose P. camembertii was grown
on minimal medium (per liter: 5 g glucose, 3.75 g KH2PO4, 0.5 g MgSO4, 0.1 g NaCl, 0.1 g
CaCl2, 0.75 g KOH, 1.2 g KNO3, 15 g agar) for 5 days at 25 C and transferred to minimal
medium containing either L-[5-3H]-tryptophan (at a concentration of 30 uCi/ml with a
specific activity of 31.5 Ci/mmol) or [14C]-acetic acid, sodium salt (at a concentration of 10
mCi/ml with a specific activity of 59 mCi/mmol), and grown at 25 C for 5 days. After that
time the same amount of cell material either from the mutant strain or from the wild-type
strain was recovered from a colony, transferred to a microcentrifuge tube and treated with
1 ml chloroform for 5 min in a microcentrifuge shaker. The mycelium was discarded and
the chloroform evaporated in a vacuum centrifuge. The residue was redissolved in 10 ul
chloroform and applied to a thin layer chromatography plate (Kiesel-gel 60, Merck,
Darmstadt) and chromatographed in two dimensions (solvent systems: first dimension,
chloroform/methanol:9/1; second dimension chloroform/isobutylmethylketone:4/1). The
Rf values were determined by dividing the migration length of the respective spot by the
migration length of the solvent. After chromatographic separation the plates were
autoradiographed.
The mutant and the wild-type strain were first labelled with 3H-tryptophan. If there
is no regulatory feedback mechanism to prevent an accumulation of the respective precursor
there should be an accumulation of a new metabolite just "in front" of the mutation if the
mutation is located within the biosynthetic branch between tryptophan and CA. The results
of the experiment are shown in Figure 1. Beside the clear signal of CA in the case of the
wild type, there was no other chloroform extractable 3H-tryptophan-labelled material under
the conditions used. In the case of the mutant strain, there was no signal at all, indicating
the mutant had not integrated radioactively labelled tryptophan into extractable precursors
of CA. These results indicate the mutation is probably not located within this biosynthetic
pathway, assuming there is no regulatory feedback mechanism. The fact that the mutant
strain was also able to grow on minimal medium without tryptophan indicates the mutation
has not affected the pathway for biosynthesis of this amino acid.
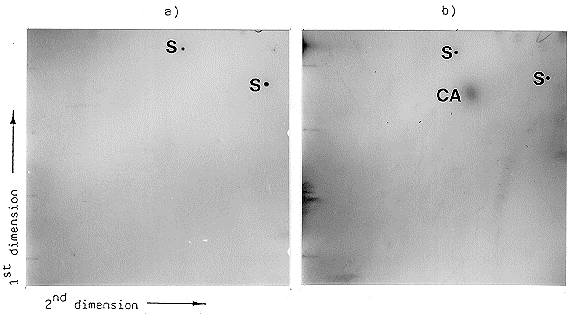
Figure 1. Autoradiography of the two-dimensional thin layer chromatography of the
chloroform-extractable metabolites of the [3H]tryptophan-labelled mutant (a) and the wild
type (b). The extraction of CA and the solvent systems used in each dimension are
described in the text. For two-dimensional thin layer chromatography, baselines for each
dimension were marked on the thin layer plate. The two baselines formed a right angle and
at the crossing point the extract was loaded. Two spots of pure CA were applied as
standard samples on the other ends of the two baselines. In the first dimension the plate
was chromatographed just below the application point of the standard sample for the second
dimension and vice versa. The localization of pure CA used as control in each dimension
was marked with 0.1 uCi of [3H] tryptophan.
Standard samples = s, extracted cyclopiazonic acid = CA
To test the possibility that the mutation has influenced the pathway for isoprenoid
biosynthesis, the mutant and the wild-type strain were grown on minimal medium containing
14C-acetate. Acetate is the precursor of the isoprenoids, so labelling with 14C-acetate should
give different patterns of extractable metabolites if the mutation is located within this
pathway. Figure 2 shows the result of this experiment. There is a different signal pattern
between the mutant (Fig. 2a) and the wild type (Fig. 2b). In the case of the mutant there
are clearly differences in the separation of the spots on the baseline of the first dimension.
These spots are not able to migrate by using the second solvent system. A clear resolution
of one spot in the second dimension at a higher Rf value (0.868) could be observed in the
case of the mutant (Fig. 2a, 1). In the case of the wild type, resolution of two spots
occurred (Fig. 2b, 2 and 3). The solvent system used in the second dimension shows poor
separation capacity for these substances. The fact that the Rf values are different in the
mutant compared with the wild type (0.723 and 0.568) indicates different labelled
substances. The results show that the mutation affects different chloroform extractable
metabolites labelled with 14C-acetate rather than affecting a single substance. Similar
complex changes of the labelling pattern of the mutant compared to that of the wild type
could be observed when the strains were incubated for a longer period of time (14 days or
21 days) on medium containing 14C-acetate (data not shown). In these cases the overall
patterns from both the mutant and the wild type were different from the patterns seen in
Fig. 2, indicating a change of the accumulated labelled metabolites during the time course
of incubation. These results show that the mutation leads to complex changes in the pattern
of the 14C-acetate-labelled metabolites and suggests it is located in the pathway for
isoprenoid biosynthesis.
Alternative possibilities that condensation of acetyl-CoA residues to tryptophan is
abolished or that the mutation affected the pathway from tryptophan to CA, and that an
accumulation of a precursor of that pathway is prevented by a feedback mechanism are not
likely because in any case the 14C-acetate labelling pattern should not be changed.
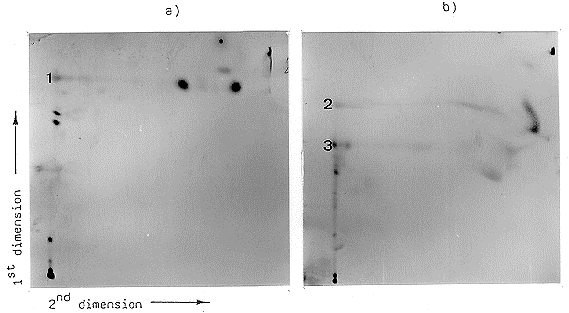
Figure 2. Autoradiography of the two-dimensional thin layer chromatography of the chloroform-extractable
metabolites of the [14C]acetate-labelled mutant (a) and the wild-type strain (b). Extraction of CA and the solvent system of each dimension are described in the text. 1,2,3 = separable spots by using the second solvent system.

