Mitotic instability in benomyl-resistant transformants of a fluffy strain of Neurospora
crassa
Claude Rossier, G. Brazil and Anne Utz-Pugin - Laboratory of General Microbiology,
Department of Botany and Plant Biology, University of Geneva, Sciences III, 30 Quai
Ernest-Ansermet, CH-1211 Geneva 4, Switzerland
The isolation of the beta-tubulin gene from a benomyl-resistant Neurospora crassa strain
(Orbach et al. 1986 Mol. Cell. Biol. 6:2452-2461) has provided a dominant selectable marker
usable in transformation experiments with N. crassa.
In order to determine whether plasmid pBT6 (bearing the benomyl-resistant beta-tubulin
gene on the pUC12 vector) stably integrated into genomic DNA, we used it to transform
the morphological mutant fluffy of N. crassa (FGSC 45). This strain can be conveniently
used in transformation experiments because it generates only uninucleate microconidia,
allowing the isolation of homokaryons and avoiding the complications in isolation via
ascospores (Rossier et al. 1985 Curr. Genet. 10:313-320). The fate of pBT6 DNA was
analyzed in transformants grown under selective or non-selective conditions.
To produce microconidia, the fluffy strain was grown for 7 days at 25 C in Petri
plates on Westergaard and Mitchell's synthetic medium supplemented with 1% acetate as
sole carbon source (acetate medium). Transformation experiments were performed
according to Orbach et al. (1986 Mol. Cell. Biol. 6:2452-2461) with modifications.
Microconidia were inoculated in Neurospora minimal medium (Difco no. 0817-01) at a
concentration of 1 x 10(7) cells per ml. Cultures were incubated 24 hours at 33 C at 150 rpm.
Germinating microconidia (about 5 x 10(8)) were washed 3 times with distilled water at room
temperature and resuspended in 10 ml 1 M sorbitol to which 2 ml Novozym 234 (Novo
Industri A/S, batch 1199, 5 mg/ml in 1 M sorbitol) were added. Digestion was performed
for 1 hour at 30 C at 100 rpm. After centrifugation, spheroplasts were washed twice with
10 ml 1 M sorbitol and once with 10 ml 5 mM Tris-HCl buffer pH 8.0, 1 M sorbitol, 50 mM
CaCl2 (transformation buffer). Spheroplasts in transformation buffer were diluted with 0.6
volume distilled water just before transformation (Akins and Lambowitz 1985 Mol. Cell.
Biol. 5:2272-2278). Five microliters of DMSO and 0.1 ml 50 mM Tris-HCl pH 8.0
containing 40% polyethylene glycol 4000 (PEG) (BDH) and 50 mM CaCl2 were then added
to the spheroplasts in 0.4 ml diluted transformation buffer. Ten micrograms of plasmid
pBT6 DNA purified twice by CsCl ultracentrifugation in 100 ul TE, pH 8.0, and
preincubated 10 min with 25 ul heparin solution (5mg/ml, freshly prepared in
transformation buffer), were then added to the transformation mixture. After 30 min
incubation on ice, 5 ml PEG-Tris-CaCl2 solution were added and incubation continued for
20 min at room temperature. Appropriate aliquots of the transformation mixture were
inoculated with 3% agar, 1 M sorbitol, 2% sorbose, 0.05% fructose and 0.05% glucose) at
45 C. This medium was poured on a 25 ml bottom layer (Vogel's medium N containing 2%
sorbose, 0.05% fructose, 0.05% glucose and 1.5% agar to which 0.5 ug/ml benomyl was
added after autoclaving). Plates were incubated for 3 days at 33 C.
Spheroplasts derived from microconidia germinated for 24 hours could be
transformed with pBT6 with a frequency of 1000 to 4000 transformants per ug DNA.
Microconidia incubated for 12 h were still ungerminated and were not suited for
transformation due to poor formation of spheroplasts. Homokaryotic derivatives from the
presumably heterokaryotic primary transformants were obtained by isolating colonies grown
from microconidia inoculated on selective (0.25 ug/ml benomyl) Vogel's medium N
supplemented with 2% sorbose, 0.05% fructose and 0.05% glucose.
The proportion of transformed nuclei and their stability in benomyl-resistant
transformants were determined by genetic analysis of microconidia produced on selective
(0.5 ug/ml benomyl) or non-selective acetate medium, assuming that the microconidia
contain a random sample of the nuclei present in the mycelium. The percentage of
transformed microconidia was determined by counting the colonies obtained from an equal
number of microconidia plated onto selective (0.25 ug/ml benomyl) and non-selective
Vogel's medium N (both contained 2% agar plus 2% sorbose, 0.05% glucose and 0.05%
fructose). A variable degree of mitotic instability was observed during growth in the
absence of selective pressure. Under such growth conditions, the microconidia of some
transformants kept the transformed character (35% of benomyl resistant microconidia in
transformant 1f) while microconidia of others lost it (<0.3% of transformed microconidia
in transformant 3i).
To investigate at the molecular level the effects of the suppression of selective
pressure on the fate of the transforming DNA, 3 transformants (1f, 9 and 3i) were grown
in Vogel's medium N plus (0.25 ug/ml) or minus benomyl. DNA was then extracted and
purified by CsCl ultracentrifugation. Undigested plasmid pBT6 DNA and undigested
genomic DNAs from the transformants grown in the presence of benomyl were
electrophoresed on 0.8% agarose gels and probed with nick-translated pBT6. No bands
were detected that would correspond to the relaxed circular or supercoiled configurations
of the plasmid (results not shown). This is consistent with the integration of pBT6 into
genomic DNA and with the absence of free, autonomously replicating plasmids.
ClaI does not cut pBT6. In
untransformed fluffy, pBT6 hybridized
to a single ClaI DNA fragment of
about 13 kb bearing the resident beta-tubulin gene (Fig. 1). In all genomic
DNAs from the transformants, pBT6
hybridized to this band and to another
ClaI fragment of higher molecular
weight indicating that integration
occurred by non-homologous
recombination. However, the ClaI
fragments bearing pBT6 sequences
were shorter than the corresponding
undigested DNAs, ruling out the
presence of non-integrated circular
oligomers of pBT6 (date not shown).
In the absence of selective pressure,
pBT6 sequences were maintained
(transformant 1f) or lost (transformant
3i). In transformant 9, the intensity on
the autoradiogram of the ClaI
fragment bearing the pBT6 sequences
is weaker after growth in the absence
of selective pressure (Fig. 1). This
suggests that some nuclei lost the
pBT6 sequences, in agreement with
the finding that only 7% of
transformant 9 microconidia kept the
transformed character under such
growth conditions, as determined by
genetic analysis.
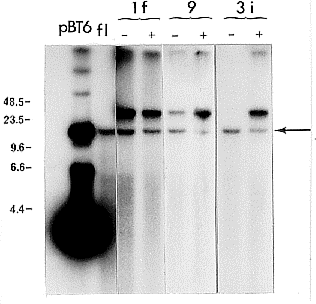
Figure 1. Genomic Southern blot analysis of DNAs isolated from untransformed strain
fluffy (fl) of N. crassa and from 3 benomyl-resistant transformants (1f, 9 and 3i).
Transformants were grown in the absence of selective pressure (-) or in the presence (+)
of 0.25 µg/ml benomyl. 1 ug of DNA from each strain and 1 ng of plasmid pBT6 were
electrophoresed after digestion with ClaI on 0.8% agarose gels and probed with
32P-labelled
pBT6. As ClaI does not cut pBT6, it migrated mainly in the supercoiled configuration.
The
arrow points to the 13 kb ClaI DNA fragment bearing the resident beta-tubulin gene. Size
markers ( lambda DNA digested with HindIII) are shown in kb; fl: untransformed fluffy strain.
These results strongly suggest
that mitotic instability observed in the absence of selective pressure is due to excision of
integrated plasmid sequences. These observations contrast with the mitotic stability
observed in N. crassa with other transformation systems (Case 1986 Genetics 113:569-587;
Avalos et al. 1989 Curr. Genet. 16:369-372) and with the finding that pBT6 gave rise to
mitotically stable transformants in Podospora anserina (Fernandez-Larrea and Stahl 1989
Curr. Genet. 16:57-60).
Introduction of DNA into N. crassa can lead to sequence instability in the sexual
phase of the life cycle (RIP process, Selker et al. 1987 Cell 51:741-752). Our results suggest
that another mechanism operating during vegetative growth serves to preserve the
organization of the N. crassa genome.
Acknowledgements: I would like to acknowledge the support of Gilbert Turian. This work
was supported by the Swiss National Science Foundation (Grant no. 3.073-0.84)
