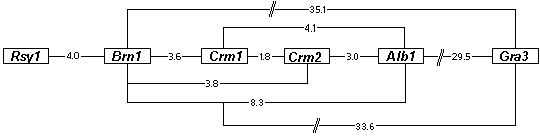
Figure 1. Proposed map of the gene cluster encoding melanin biosynthetic enzymes on chromosome 1 of C. heterostrophus, based on the two point cross data in Table 1. The gene order and the map distances must be considered tentative, since two point data may not accurately reflect recombination frequencies and do not prove gene order. Note that Rsy1 could be on either side of Brn1 and that linkage of Gra3 to the cluster is tenuous because of the distance between them. Numbers indicate cM.
In addition to the randomly isolated ascospores reported in Table 1, a few complete
tetrads (four sets of twins/ascus) were recovered from each cross; they were predominantly
parental ditypes with at least one tetratype ascus from each pair of parents (nonparental
ditypes were not found in any progeny). Color patterns in tetratype asci indicated the
following epistatic relationships [evidence for epistasis of brn1 over rsy1 is based on the
relative high abundance of brn1 progeny from the brn1 X rsy1 cross (Table 1), not on
tetratype asci]:
alb1 > crm1 > crm2 > brn1 > rsy1 > gra3
Table 1. Progeny tests of two point crosses between C. heterostrophus color mutants.a Recombinant progeny included wild types (recognizable by their dark brown-green color) and double mutants (which could not be recognized because they expressed the color of the epistatic parent). The number of double mutants in each progeny was assumed to equal that of wild types, and the wild type number was doubled to calculate the recombination frequency.
Cross Number of progeny Map distancea _________________ _____________________________________ Parent 1 Parent 2 Parent 1 type Parent 2 type Wild type (cM) ______________________________________________________________________________ brn1 alb1 47 45 4 8.3 brn1 crm1 75 86 3 3.6 brn1 crm2 76 80 3 3.8 alb1 crm1 74 70 3 4.1 alb1 crm2 97 97 3 3.0 crm1 crm2 612 513 10 1.8 gra3 alb1 58 92 26 29.5 gra3 brn1 27 34 13 35.1 gra3 crm1 24 55 16 33.6 rsy1 brn1 87 108 4 4.0 ______________________________________________________________________________
An attempt was made to test the gene order shown in Figure 1 by scoring progeny
from the three point cross alb1;brn1 X crm2. Of 945 progeny isolated, 478 were alb1, 431
were crm2, 24 were brn1, and 12 were wild type. These data are not consistent with the
gene order Brn1-Alb1-Crm2, which would require that the Brn phenotype represent double
cross overs as the least frequent class, which it is not. However, because epistasis obscured
certain recombinant classes of progeny, the data do not distinguish between the other two
possible gene orders: Brn1-Crm2-Alb1 (which the two point data in Table 1 suggest) and
Crm2-Brn1-Alb1.
Although albino (alb1) strains of C. heterostrophus cannot survive in the field, they cause lesions qualitatively similar to those of wild type on the host plant (corn) in a growth chamber or greenhouse (Fry et al. 1984 Phytopathology 74:175-178). To determine if there are quantitative differences in virulence among color mutants or between color mutants and wild type, lesions on corn plants (in a greenhouse) caused by strains carrying alb1, crm1, crm2, brn1, or rsy1 were counted and measured, and compared with the sizes and numbers of lesions caused by wild type. No statistically significant differences were detected in any pair-wise comparison, suggesting that melanin plays no significant role in the virulence of C. heterostrophus to its host, in contrast to certain other fungi such as Colletotrichum lagenarium (Kubo et al. 1991 Mol. Plant-Microbe Int. 4:440-445) and Magnaporthe grisea (Chumley and Valent 1990 Mol. Plant-Microbe Int. 3:135-143), which require melanin for penetration of the host epidermis.