.
Cloning of the copper-inducible metallothionein (cmt) promoter from Neurospora crassa.
J. Ohrnberger and R. A. Akins, Department of Biochemistry, Wayne State University, Detroit MI
48201
There are only a limited number of vectors with inducible promoters that are convenient for
in vivo expression in Neurospora crassa. Promoters have been identified and cloned that are induced
with blue-light (bli-4; Pietschmann et al 1991 Fungal Genetics Newsl. 38:85-6) or by quinic acid (qa-2;
Campbell et al 1994 Fungal Genetics Newsl. 41:20-1). Constitutive promoters have also been used,
derived from the beta-tubulin gene bml (Nakano et al 1993 Fungal Genetics Newsl. 40:54-6). The
glucose-repressible promoter of grg-1 has also been used (Nakano ibid; Pall and Brunelli 1994 Fungal
Genetics Newsl. 41:63-4). The promoter of the N. crassa copper metallothionein gene (cmt) is
capable of induction levels of at least 100-fold (Munger et al. 1987 J. Biol. Chem. 262:7363-7) and
has been used to express tyrosinase and laccase (Kupper et al. 1990 Curr. Genet. 18:331-5; Schilling
et al. 1992 Curr. Genet. 22:197-203).
The use of the cmt promoter could enhance studies where selective expression is essential.
Unfortunately, published vectors are no longer available. Our laboratory has undertaken the
reisolation of the promoter in a convenient construct. A partial sequence of the cmt loci was
published and deposited in EMBL database (#X03009). From this, four primers were constructed
(Figure 1A) to generate PCR fragments 1 kb (primers 1 to 4), 700 bp (primers 1 to 3), and 400 bp
(primers 2 to 4) using genomic DNA from wild-type strain 74A as a template. Each fragment cross-
hybridized with the others, suggesting that each derived from overlapping chromosomal regions.
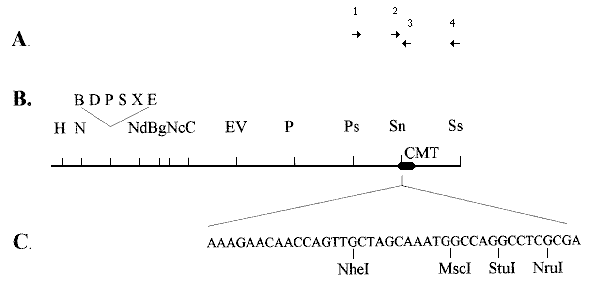
Figure 1. The copper-inducible metallothionein (cmt) gene.
A. Primers used to amplify the region
from genomic DNA. Primers, 5 ->3 : 1,GACATCACATGAACATTGCA; 2,
GGCGCTTCTTCCTGCAACTGCGGCTCTGGC ; 3, AAGATGGGATCGGACAGGCG; 4,
CGGGCGTTGTCATCACAG. Primers are positioned relative to the restriction fragment in panel
B corresponding to the relevant region of genomic DNA.
B. Restriction map of a 4.1 kb region of
cosmid G9:12G Restriction enzymes: H, HindIII; N, NotI; B, BamHI; D, DraI; P, PvuI; S, SphI; X, XhoI; E, EcoRI; Nd, NdeI; Bg, BglII; Nc, NcoI; C, ClaI; EV, EcoRV; Ps, PstI; Sn, SnaBI; Ss,
SspI.
C. Insertion of a multicloning sequence at the 3 terminus of the inducible promoter region of
the cmt gene. The region from the HindIII site to the indicated sequence was derived by PCR and
cloned back into the HindIII/EcoRV sites of Bluescript SK. Inserts can be cloned into the NheI
site to use their own ATG codon, or into the three other restriction enzyme sites to use the encoded
ATG sequence in any of the three reading frames.
The Orbach/Sachs library and the Vollmer/Yanofsky N. crassa genomic libraries (FGSC)
were screened with a random-hexamer labeled probe derived by gel-purifying the 1 kb PCR
fragment generated from primers 1 and 4 (Figure 1A). The Vollmer/Yanofsky library did not
yield any hybridizing signals, even upon repeated probing. In contrast, the Orbach/Sachs library
had six strongly hybridizing colonies. Four were isolated and the specific hybridization was
confirmed by Southern analysis. Cosmids G4:1H, G9:12G, G10:10C, and X16:10H, hybridized to
all PCR fragments recovered from the genomic PCR. Results of RFLP mapping (Metzenberg et
al. 1984 Fungal Genetics Newsl. 31: 35-42) indicated that cosmid G9:12G contains sequences from
LGVI, as would be expected of the cmt gene (Fig.2). PCR fragments were generated from this
cosmid that were identical to those deriving from genomic DNA.
RFLP type :
OMMMMMOMOOMMMMO--OOOOOOOMMOOO-MMMM-OMO
| | | |
4450 4460 4470 4480
Isolate No.
Figure 2. Sequences of cosmid G9:12G map to LGVI by RFLP. The RFLP was detected using
PstI on parental strain FGSC 4488 (O) and FGSC 2225 (M). Progeny from a cross of these two
strains, the MultiCentII kit (FGSC) were scored for the RFLP as indicated. -: not scorable; M:
Mauriceville pattern; O: Oak Ridge pattern. Underlined letters: difference in RFLP with the
closest existing probe, RAPD marker R15.7 (Metzenberg and Grotelueschuen 1994 Fungal
Genetics Newsl. 40:130-133).
A segment of cosmid G9:12G was subcloned as a 4.1 kb HindIII/SspI fragment in
Bluescript SK using the HindIII and EcoRV sites. The identity of the clone was confirmed by
sequencing into the SspI end with the primer T7/T3(Gibco-BRL). This sequence was identical
to that reported by Munger et al. (1985 EMBO J. 4:2665-8). A restriction map is provided (Figure
1B).
We have taken two approaches to contruct more useful promoter derivatives. First, a
multicloning sequence has been inserted into the full length promoter (Figure 1C). Secondly, the
entire 4 kb upstream region is reportedly required for a functional inducible promoter. It is
known that large deletions in the upstream region render the promoter constitutive (Munger et
al. 1985 EMBO J. 4:2665-8). Deletion analysis is being done from the 5 end of the current
promoter to generate smaller clones that still retain inducibility.
Preliminary experiments using the coding region of the hph gene cloned behind the full-
length promoter fragment indicated that 0.5 mM CuSO4 could induce high level resistance to
hygromycin B. These experiments were hindered by interference of the CuSO4 with hygromycin
B. A precipitate formed that raised the level of background growth and made the assay difficult
to interpret is some tests. A new construct with a different reporter is being made to confirm and
quantitate the level of inducibility of the promoter with CuSO4 and to facilitate further deletion
analysis. All constructs will be made available to the FGSC upon verification.
